Abstract
We developed a metagenomic next-generation sequencing (mNGS) test using cell-free DNA from body fluids to identify pathogens. The performance of mNGS testing of 182 body fluids from 160 patients with acute illness was evaluated using two sequencing platforms in comparison to microbiological testing using culture, 16S bacterial PCR and/or 28S–internal transcribed ribosomal gene spacer (28S–ITS) fungal PCR. Test sensitivity and specificity of detection were 79 and 91% for bacteria and 91 and 89% for fungi, respectively, by Illumina sequencing; and 75 and 81% for bacteria and 91 and 100% for fungi, respectively, by nanopore sequencing. In a case series of 12 patients with culture/PCR-negative body fluids but for whom an infectious diagnosis was ultimately established, seven (58%) were mNGS positive. Real-time computational analysis enabled pathogen identification by nanopore sequencing in a median 50-min sequencing and 6-h sample-to-answer time. Rapid mNGS testing is a promising tool for diagnosis of unknown infections from body fluids.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
Metagenomic sequencing data (FASTQ files) after removal of human genomic reads have been deposited in the NCBI Sequence Read Archive (SRA) (PRJNA558701, under umbrella project PRJNA234047).
Code availability
SURPI+ v.1.0 (https://github.com/chiulab/SURPI-plus-dist) and SURPIrt v.1.0 software (https://github.com/chiulab/SURPIrt-dist) have been deposited on GitHub and are available for download for research use only. Linux (Ubuntu 16.04.6) and Python (python 2.7.12) scripts used for construction of dual-use Illumina and nanopore barcodes are provided in the Supplementary Information, ‘Pipeline for Designing 37mer Barcodes’. Other custom scripts for ROC curve and read length analysis have been deposited on Github (https://github.com/wei2gu/2020-NGSInfectedBodyFluids/).
References
Messacar, K., Parker, S. K., Todd, J. K. & Dominguez, S. R. Implementation of rapid molecular infectious disease diagnostics: the role of diagnostic and antimicrobial stewardship. J. Clin. Microbiol. 55, 715–723 (2017).
Rea, B., Maisel, J. R., Glaser, L. & Alby, K. Identification of clinically relevant mycobacterial species after extended incubation times in the BACTEC MGIT System. Am. J. Clin. Pathol. 151, 63–67 (2019).
Janda, J. M. & Abbott, S. L. 16S rRNA gene sequencing for bacterial identification in the diagnostic laboratory: pluses, perils, and pitfalls. J. Clin. Microbiol. 45, 2761–2764 (2007).
Petti, C. A., Polage, C. R. & Schreckenberger, P. The role of 16S rRNA gene sequencing in identification of microorganisms misidentified by conventional methods. J. Clin. Microbiol. 43, 6123–6125 (2005).
Reuwer, A. Q., van den Bijllaardt, W., Murk, J. L., Buiting, A. G. M. & Verweij, J. J. Added diagnostic value of broad-range 16S PCR on periprosthetic tissue and clinical specimens from other normally sterile body sites. J. Appl. Microbiol. 126, 661–666 (2019).
Wilson, M. R. et al. Actionable diagnosis of neuroleptospirosis by next-generation sequencing. N. Engl. J. Med. 370, 2408–2417 (2014).
Wilson, M. R. et al. Clinical metagenomic sequencing for diagnosis of meningitis and encephalitis. N. Engl. J. Med. 380, 2327–2340 (2019).
Glimaker, M. et al. Adult bacterial meningitis: earlier treatment and improved outcome following guideline revision promoting prompt lumbar puncture. Clin. Infect. Dis. 60, 1162–1169 (2015).
Kumar, A. et al. Duration of hypotension before initiation of effective antimicrobial therapy is the critical determinant of survival in human septic shock. Crit. Care Med. 34, 1589–1596 (2006).
Weiss, S. L. et al. Delayed antimicrobial therapy increases mortality and organ dysfunction duration in pediatric sepsis. Crit. Care Med. 42, 2409–2417 (2014).
Llor, C. & Bjerrum, L. Antimicrobial resistance: risk associated with antibiotic overuse and initiatives to reduce the problem. Ther. Adv. Drug Saf. 5, 229–241 (2014).
Chiu, C. Y. & Miller, S. A. Clinical metagenomics. Nat. Rev. Genet. 20, 341–355 (2019).
Gu, W., Miller, S. & Chiu, C. Y. Clinical metagenomic next-generation sequencing for pathogen detection. Annu. Rev. Pathol. 14, 319–338 (2019).
Simner, P. J., Miller, S. & Carroll, K. C. Understanding the promises and hurdles of metagenomic next-generation sequencing as a diagnostic tool for infectious diseases. Clin. Infect. Dis. 66, 778–788 (2018).
Blauwkamp, T. A. et al. Analytical and clinical validation of a microbial cell-free DNA sequencing test for infectious disease. Nat. Microbiol. 4, 663–674 (2019).
Charalampous, T. et al. Nanopore metagenomics enables rapid clinical diagnosis of bacterial lower respiratory infection. Nat. Biotechnol. 37, 783–792 (2019).
Ivy, M. I. et al. Direct detection and identification of prosthetic joint infection pathogens in synovial fluid by metagenomic shotgun sequencing. J. Clin. Microbiol. 56, e00402-18 (2018).
Langelier, C. et al. Integrating host response and unbiased microbe detection for lower respiratory tract infection diagnosis in critically ill adults. Proc. Natl Acad. Sci. USA 115, E12353–E12362 (2018).
Leo, S. et al. Detection of bacterial pathogens from broncho-alveolar lavage by next-generation sequencing. Int. J. Mol. Sci. 18, 2011 (2017).
Miller, S. et al. Laboratory validation of a clinical metagenomic sequencing assay for pathogen detection in cerebrospinal fluid. Genome Res. 29, 831–842 (2019).
Schlaberg, R. et al. Validation of metagenomic next-generation sequencing tests for universal pathogen detection. Arch. Pathol. Lab Med. 141, 776–786 (2017).
Schmidt, K. et al. Identification of bacterial pathogens and antimicrobial resistance directly from clinical urines by nanopore-based metagenomic sequencing. J. Antimicrob. Chemother. 72, 104–114 (2017).
Thoendel, M. J. et al. Identification of prosthetic joint infection pathogens using a shotgun metagenomics approach. Clin. Infect. Dis. 67, 1333–1338 (2018).
Wilson, M. R. et al. Chronic meningitis investigated via metagenomic next-generation sequencing. JAMA Neurol. 75, 947–955 (2018).
Lucas, M. J., Brouwer, M. C. & van de Beek, D. Neurological sequelae of bacterial meningitis. J. Infect. 73, 18–27 (2016).
Schlaberg, R. et al. Viral pathogen detection by metagenomics and pan-viral group polymerase chain reaction in children with pneumonia lacking identifiable etiology. J. Infect. Dis. 215, 1407–1415 (2017).
Costales, C. & Butler-Wu, S. M. A real pain: diagnostic quandaries and septic arthritis. J. Clin. Microbiol. 56, e01358-17 (2018).
McGill, F., Heyderman, R. S., Panagiotou, S., Tunkel, A. R. & Solomon, T. Acute bacterial meningitis in adults. Lancet 388, 3036–3047 (2016).
Singal, A. K., Salameh, H. & Kamath, P. S. Prevalence and in-hospital mortality trends of infections among patients with cirrhosis: a nationwide study of hospitalised patients in the United States. Aliment. Pharm. Ther. 40, 105–112 (2014).
Greninger, A. L. et al. Rapid metagenomic identification of viral pathogens in clinical samples by real-time nanopore sequencing analysis. Genome Med. 7, 99 (2015).
Faria, N. R. et al. Mobile real-time surveillance of Zika virus in Brazil. Genome Med. 8, 97 (2016).
Kafetzopoulou, L. E. et al. Metagenomic sequencing at the epicenter of the Nigeria 2018 Lassa fever outbreak. Science 363, 74–77 (2019).
Quick, J. et al. Real-time, portable genome sequencing for Ebola surveillance. Nature 530, 228–232 (2016).
Raith, E. P. et al. Prognostic accuracy of the SOFA Score, SIRS Criteria, and qSOFA score for in-hospital mortality among adults with suspected infection admitted to the intensive care unit. JAMA 317, 290–300 (2017).
Deng, X. et al. Metagenomic sequencing with spiked primer enrichment for viral diagnostics and surveillance. Nat. Microbiol. 5, 443–454 (2020).
Fan, H. C., Blumenfeld, Y. J., Chitkara, U., Hudgins, L. & Quake, S. R. Analysis of the size distributions of fetal and maternal cell-free DNA by paired-end sequencing. Clin. Chem. 56, 1279–1286 (2010).
De Vlaminck, I. et al. Temporal response of the human virome to immunosuppression and antiviral therapy. Cell 155, 1178–1187 (2013).
Focosi, D., Antonelli, G., Pistello, M. & Maggi, F. Torquetenovirus: the human virome from bench to bedside. Clin. Microbiol. Infect. 22, 589–593 (2016).
Abril, M. K. et al. Diagnosis of Capnocytophaga canimorsus sepsis by whole-genome next-generation sequencing. Open Forum Infect. Dis. 3, ofw144 (2016).
Grumaz, S. et al. Next-generation sequencing diagnostics of bacteremia in septic patients. Genome Med. 8, 73 (2016).
Long, Y. et al. Diagnosis of sepsis with cell-free DNA by next-generation sequencing technology in ICU patients. Arch. Med. Res. 47, 365–371 (2016).
De Mattos-Arruda, L. et al. Cerebrospinal fluid-derived circulating tumour DNA better represents the genomic alterations of brain tumours than plasma. Nat. Commun. 6, 8839 (2015).
Dudley, J. C. et al. Detection and surveillance of bladder cancer using urine tumor DNA. Cancer Discov. 9, 500–509 (2019).
Pan, W., Gu, W., Nagpal, S., Gephart, M. H. & Quake, S. R. Brain tumor mutations detected in cerebral spinal fluid. Clin. Chem. 61, 514–522 (2015).
Springer, S. U. et al. Non-invasive detection of urothelial cancer through the analysis of driver gene mutations and aneuploidy. eLife 7, e32143 (2018).
Wang, Y. et al. Evaluation of liquid from the Papanicolaou test and other liquid biopsies for the detection of endometrial and ovarian cancers. Sci. Transl. Med. 10, eaap8793 (2018).
Corless, C. E. et al. Contamination and sensitivity issues with a real-time universal 16S rRNA PCR. J. Clin. Microbiol. 38, 1747–1752 (2000).
Klindworth, A. et al. Evaluation of general 16S ribosomal RNA gene PCR primers for classical and next-generation sequencing-based diversity studies. Nucleic Acids Res. 41, e1 (2013).
Salipante, S. J. et al. Performance comparison of Illumina and ion torrent next-generation sequencing platforms for 16S rRNA-based bacterial community profiling. Appl. Environ. Microbiol. 80, 7583–7591 (2014).
Wang, Y. & Qian, P. Y. Conservative fragments in bacterial 16S rRNA genes and primer design for 16S ribosomal DNA amplicons in metagenomic studies. PLoS ONE 4, e7401 (2009).
Hoggard, M. et al. Characterizing the human Mycobiota: a comparison of small subunit rRNA, ITS1, ITS2, and large subunit rRNA genomic targets. Front. Microbiol. 9, 2208 (2018).
Chan, K. C. et al. Molecular characterization of circulating EBV DNA in the plasma of nasopharyngeal carcinoma and lymphoma patients. Cancer Res. 63, 2028–2032 (2003).
Greninger, A. L. et al. Clinical metagenomic identification of Balamuthia mandrillaris encephalitis and assembly of the draft genome: the continuing case for reference genome sequencing. Genome Med. 7, 113 (2015).
Mongkolrattanothai, K. et al. Neurobrucellosis: unexpected answer from metagenomic next-generation sequencing. J. Pediatr. Infect. Dis. 6, 393–398 (2017).
Sardi, S. I. et al. Coinfections of Zika and Chikungunya viruses in Bahia, Brazil, identified by metagenomic next-generation sequencing. J. Clin. Microbiol. 54, 2348–2353 (2016).
Wilson, M. R. et al. Diagnosing Balamuthia mandrillaris encephalitis with metagenomic deep sequencing. Ann. Neurol. 78, 722–730 (2015).
World Medical Association World Medical Association Declaration of Helsinki: ethical principles for medical research involving human subjects. JAMA 310, 2191–2194 (2013).
Naccache, S. N. et al. A cloud-compatible bioinformatics pipeline for ultrarapid pathogen identification from next-generation sequencing of clinical samples. Genome Res. 24, 1180–1192 (2014).
Zaharia, M. et al. Faster and more accurate sequence alignment with SNAP. Preprint at https://arxiv.org/abs/1111.5572 (2011).
Zinter, M. S., Mayday, M. Y., Ryckman, K. K., Jelliffe-Pawlowski, L. L. & DeRisi, J. L. Towards precision quantification of contamination in metagenomic sequencing experiments. Microbiome 7, 62 (2019).
McIntyre, A. B. R. et al. Comprehensive benchmarking and ensemble approaches for metagenomic classifiers. Genome Biol. 18, 182 (2017).
Strain, M. C. et al. Highly precise measurement of HIV DNA by droplet digital PCR. PLoS ONE 8, e55943 (2013).
Taylor, S. C., Laperriere, G. & Germain, H. Droplet digital PCR versus qPCR for gene expression analysis with low abundant targets: from variable nonsense to publication quality data. Sci. Rep. 7, 2409 (2017).
Altschul, S. F., Gish, W., Miller, W., Myers, E. W. & Lipman, D. J. Basic local alignment search tool. J. Mol. Biol. 215, 403–410 (1990).
Burnham, P. et al. Urinary cell-free DNA is a versatile analyte for monitoring infections of the urinary tract. Nat. Commun. 9, 2412 (2018).
Acknowledgements
We thank H. Reyes, A. Chan, D. Ingebrigtsen, L. Dang, W. Lorizio, J. Streithorst, B. Fung and the UCSF clinical laboratory staff for assistance with orthogonal testing. We thank R. Da for assistance with sequencing runs. We thank S. Shiboski for biostatistics feedback. We thank members of the Chiu, Miller and DeRisi laboratories for feedback and support. This work was funded in part by an NIH grant (no. K08-CA230156) and a Burroughs-Wellcome Award to W.G., by Abbott Laboratories (C.Y.C.), a NIH/NHLBI grant (no. R01-HL105704, to C.Y.C.), a NIH/NIAID grant (no. R33-AI120977, to C.Y.C.), the California Initiative to Advance Precision Medicine (C.Y.C.), a UC Center for Accelerated Innovation grant funded by NIH grant U54HL119893 and NIH/NCATS UCSF-CTSI grant UL1TR000004 (C.Y.C.), and the Charles and Helen Schwab Foundation (C.Y.C.).
Author information
Authors and Affiliations
Contributions
W.G., X.D. and C.Y.C. conceived and designed the study. W.G., X.D. and C.Y.C. coordinated the study. W.G., X.D., M.L., Y.D.S., S.A., A.G., K.R., G.Y., B.B., E.D.C., C.W. and E.H. performed experiments. W.G., X.D., M.L., E.H. and C.Y.C. analyzed data. W.G., G.I., E.H. and C.Y.C. reviewed patient electronic medical records. S.F. and D.S. wrote software and performed SURPI bioinformatics analysis of mNGS data. K.Z., H.S. and G.I. enrolled patients in the study and assisted in patient data collection. A.B., M.R.W., S.M., J.L.D. and C.Y.C. provided clinical samples and resources. W.G., X.D., M.L. and C.Y.C. wrote and edited the manuscript. W.G. and C.Y.C. designed the figures. C.Y.C. supervised the study. All authors read the manuscript and agree to its contents.
Corresponding author
Ethics declarations
Competing interests
C.Y.C. is the director of the UCSF-Abbott Viral Diagnostics and Discovery Center and receives research support funding from Abbott Laboratories, Inc. C.Y.C., D.S., S.F. and S.M. are inventors on a patent application on algorithms related to SURPI+ software (no. PCT/US/16/52912, ‘Pathogen Detection using Next-Generation Sequencing’). C.Y.C., X.D. and S.F. are inventors on a patent pending on algorithms related to SURPIrt software (Case no. SF2015-154, ’Methods for Real-Time Sequencing Analysis of Infectious Diseases’). Other coauthors declare no competing interests.
Additional information
Peer review information Alison Farrell is the primary editor on this article and managed its editorial process and peer review in collaboration with the rest of the editorial team.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
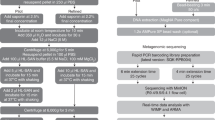
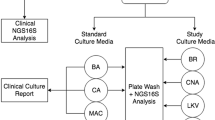
Extended Data Fig. 1 Metagenomic sequencing of body fluids.
a, Nanopore time to detection (minutes) across different body fluid types. Each data point represents the time to detection of the organism, if any, in each body fluid sample. b, Nanopore time to detection (minutes) in relation to pathogen DNA abundance in samples (reads per million, RPM). All box plots represent the median (centre), the interquartiles (minima and maxima), and 1.5 x interquartile range (whiskers). c, Precision-recall curves for Illumina and nanopore training fungal datasets. d, Precision-recall curves based on the Illumina training bacterial dataset in comparison with the composite standard. e, Precision-recall curves based on nanopore bacterial training datasets. f, Pie chart showing distribution of bacterial pathogen titers as estimated by semi-quantitative culture. g, Plot of nRPM values versus semi-quantitative bacterial titers. The nRPM corresponding to bacteria cultured in enrichment broth was significantly lower than the other higher-titer cultures (p = 0.006). h, Relative pathogen burden in positive and negative (non-infectious) body fluid samples. i, Log scale plot of the bacterium Achromobacter xylosoxidans from mNGS data, a common background contaminant in sequencing libraries. There is a log-linear relationship between the qPCR cycle threshold (Ct) value and the RPM corresponding to Achromobacter xylosoxidans. The background level of Achromobacter xylosoxidans is inversely correlated with the input concentration and is relatively constant.
Extended Data Fig. 2 ROC curves of mNGS test performance.
ROC curves are plotted from validation set data based on a clinical gold standard or composite standard. Data are presented as median true positive rates± the 95% confidence intervals. The 95% confidence interval was obtained via a bootstrap method with 2000 resampling iterations. a, Illumina dataset, bacterial detection (n = 127 samples). b, Nanopore dataset, bacterial detection (n = 43 samples), c, Illumina dataset, fungal detection (n = 127, 32 fungal organisms). d, Nanopore dataset, fungal detection (n = 43, 11 fungal organisms).
Extended Data Fig. 3 Relationship of external positive control organism titer with mNGS detection signal (expressed in nRPM).
Simple linear regression of normalized reads per million (nRPM) over four replicates per dilution factor, calculated as genome equivalents per mL (GE/mL) for a, Streptococcus uberis, b, Rhodobacter sphaeroides, c, Aspergillus oryzae, and d, Millerozyma farinosa.
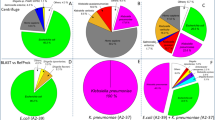
Extended Data Fig. 4 Orthogonal testing for Case S31: Klebsiella pneumoniae infection of pleural fluid.
a, Genomic coverage of K. pneumoniae from Illumina mNGS. Sequencing spanned 36,490 base pairs, or 0.65% of the K. pneumoniae genome. b, Orthogonal confirmation of K. pneumoniae by dPCR of the sequencing library. Nine negative controls from other cases were run in parallel. Out of 10 sequencing libraries, only Case S31 had any positive droplets (n = 43 of 12022 total droplets as circled). c, Orthogonal confirmation of K. pneumoniae by dPCR of the DNA extract. Three positive droplets were detected, indicating a low positive result. d, Orthogonal confirmation of K. pneumoniae by dPCR of contralateral pleural fluid (sample C31). 29 and 24 positive droplets were detected out of 2 replicates. Digital PCR targeting Streptococcus mitis on both pleural fluids did not yield any positive droplets. The positive controls for these experiments were from sheared DNA from Klebsiella pneumoniae and Streptococcus mitis respectively, whereas the negative control was water. e, Sanger sequencing of the K. pneumoniae amplicon from dPCR. Shown are sequencing traces confirming the presence of K. pneumoniae.
Extended Data Fig. 5 Orthogonal testing for Cases S88: Klebsiella aerogenes from cerebrospinal fluid and S87: Bartonella henselae from a skin abscess.
a, Orthogonal confirmation of K. aerogenes by dPCR of the DNA extract. The sample was run in parallel with 9 negative controls. Out of 10 sequencing libraries, only Case S88 had positive dPCR droplets (n = 61). b, Genomic coverage of K. aerogenes from Illumina mNGS. The assembled genomic regions spanned 536,461 bp, or 9.9% of the bacterial genome. c, Orthogonal confirmation of Bartonella henselae by dPCR of the DNA extract. Positive dPCR droplets (n = 12) are seen in abscess fluid and the positive control consisting of sheared DNA from Bartonella henselae (ATCC 49882). The negative control was water.
Extended Data Fig. 6 Length distributions of pathogen cfDNA in mNGS data.
Analysis is performed on the 87 body fluid samples sequenced on both Illumina and nanopore platforms. a, Diagram showing how original genomic DNA lengths are recovered. Paired-end sequencing data is aligned to either a human or microbial genome, followed by determination of fragment length from the start and end positions and construction of a read length histogram. b, Histogram of average DNA lengths for human, bacterial, and fungal organisms obtained from mNGS data. Human DNA is observed to peak at the stereotypical 160 bp nucleosome footprint; both bacterial and fungal DNA are most abundant at sizes of <100 bp, but a higher molecular weight tail is observed extending to 500–600 bp. c, Histogram of bacterial read lengths according to sequencing platform. Illumina and nanopore sequencing platforms produce different size distributions. d, Length analysis of mNGS reads for samples with 16S PCR performed. Comparison of the length profiles of the 16S discordant bacteria cases (S31 and S88), 16S concordant bacteria cases (mean of S10, S36, S41, S69, S85, S128), and all bacteria mean. The pathogen cfDNA in cases S31 and S88 are more fragmented, with the vast majority of fragments <300 bp. The relative paucity of longer fragments could hinder 16S PCR amplification.
Extended Data Fig. 7 Comparison of different threshold variables on the training set to calibrate the thresholds for each variable used.
The final thresholds used are circled in each ROC chart. a, Comparison of different minimal read thresholds for bacteria calling. Based on this data and prior selection of minimal reads, we selected a minimal of 3 reads for the validation set. b, Comparison of using or not using a PCR Ct value normalization for bacteria calling. Normalization resulted in higher specificity and was used on the validation set. c, Comparison of using a same-genus/same-family filter to decrease an informatics artifact where a pathogen burden is high and related species would appear at significant lower values. Using this filter improved specificity. d, Comparison of different minimal read thresholds for fungal calling. We selected a minimal of 1 read based on the significantly higher sensitivity at the lowest threshold. e, Comparison of using or not using a PCR Ct value normalization for fungal calling. Normalization resulted in higher specificity and was used on the validation set.
Supplementary information
Supplementary Information
Supplementary Tables 1–13 and discussion.
Supplementary Table 1
Clinical characteristics and mNGS results of all cases used in the accuracy study.
Supplementary Table 2
Positive and negative external controls.
Supplementary Table 3
Differences between clinical and composite standards.
Supplementary Table 4
Bacterial false positives and false negatives using Illumina and nanopore sequencing.
Supplementary Table 5
Fungal true positives, false positives and false negatives using Illumina and nanopore sequencing.
Supplementary Table 11
PCR primers and indexes used for sequencing library barcoding.
Rights and permissions
About this article
Cite this article
Gu, W., Deng, X., Lee, M. et al. Rapid pathogen detection by metagenomic next-generation sequencing of infected body fluids. Nat Med 27, 115–124 (2021). https://doi.org/10.1038/s41591-020-1105-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41591-020-1105-z
This article is cited by
-
Metagenomic next-generation sequencing of plasma cell-free DNA improves the early diagnosis of suspected infections
BMC Infectious Diseases (2024)
-
Antimicrobial resistance prediction by clinical metagenomics in pediatric severe pneumonia patients
Annals of Clinical Microbiology and Antimicrobials (2024)
-
Development and proof-of-concept demonstration of a clinical metagenomics method for the rapid detection of bloodstream infection
BMC Medical Genomics (2024)
-
Clinical application of bronchoalveolar lavage fluid metagenomics next-generation sequencing in cancer patients with severe pneumonia
Respiratory Research (2024)
-
Diagnostic strategy of metagenomic next-generation sequencing for gram negative bacteria in respiratory infections
Annals of Clinical Microbiology and Antimicrobials (2024)