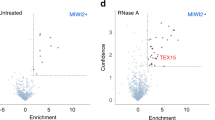
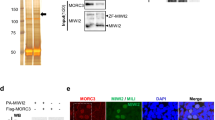
Abstract
In mammals, the acquisition of the germline from the soma provides the germline with an essential challenge: the need to erase and reset genomic methylation1. In the male germline, RNA-directed DNA methylation silences young, active transposable elements2,3,4. The PIWI protein MIWI2 (PIWIL4) and its associated PIWI-interacting RNAs (piRNAs) instruct DNA methylation of transposable elements3,5. piRNAs are proposed to tether MIWI2 to nascent transposable element transcripts; however, the mechanism by which MIWI2 directs the de novo methylation of transposable elements is poorly understood, although central to the immortality of the germline. Here we define the interactome of MIWI2 in mouse fetal gonocytes undergoing de novo genome methylation and identify a previously unknown MIWI2-associated factor, SPOCD1, that is essential for the methylation and silencing of young transposable elements. The loss of Spocd1 in mice results in male-specific infertility but does not affect either piRNA biogenesis or the localization of MIWI2 to the nucleus. SPOCD1 is a nuclear protein whose expression is restricted to the period of de novo genome methylation. It co-purifies in vivo with DNMT3L and DNMT3A, components of the de novo methylation machinery, as well as with constituents of the NURD and BAF chromatin remodelling complexes. We propose a model whereby tethering of MIWI2 to a nascent transposable element transcript recruits repressive chromatin remodelling activities and the de novo methylation apparatus through SPOCD1. In summary, we have identified a previously unrecognized and essential executor of mammalian piRNA-directed DNA methylation.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
All mRNA expression data that support the findings of this study have been deposited at Array Express under accession number E-MTAB-7985. The Methyl-seq data generated in this study have been deposited at ArrayExpress under the accession number E-MTAB-7997. The sRNA-seq and RNA-seq data generated in this study have been deposited at the Gene Expression Omnibus under the accession number GSE131377. Data for the IP-MS experiments were deposited at ProteomeXchange under the accession number PXD016701. Source data are provided with this paper.
Code availability
Scripts used for the Methyl-seq, RNA-seq and sRNA-seq analysis are available on github (https://github.com/rberrens/SPOCD1-piRNA_directed_DNA_met).
References
Tang, W. W., Kobayashi, T., Irie, N., Dietmann, S. & Surani, M. A. Specification and epigenetic programming of the human germ line. Nat. Rev. Genet. 17, 585–600 (2016).
Aravin, A. A., Sachidanandam, R., Girard, A., Fejes-Toth, K. & Hannon, G. J. Developmentally regulated piRNA clusters implicate MILI in transposon control. Science 316, 744–747 (2007).
Carmell, M. A. et al. MIWI2 is essential for spermatogenesis and repression of transposons in the mouse male germline. Dev. Cell 12, 503–514 (2007).
Molaro, A. et al. Two waves of de novo methylation during mouse germ cell development. Genes Dev. 28, 1544–1549 (2014).
Kuramochi-Miyagawa, S. et al. DNA methylation of retrotransposon genes is regulated by Piwi family members MILI and MIWI2 in murine fetal testes. Genes Dev. 22, 908–917 (2008).
Walsh, C. P., Chaillet, J. R. & Bestor, T. H. Transcription of IAP endogenous retroviruses is constrained by cytosine methylation. Nat. Genet. 20, 116–117 (1998).
Ohinata, Y. et al. Blimp1 is a critical determinant of the germ cell lineage in mice. Nature 436, 207–213 (2005).
Chedin, F., Lieber, M. R. & Hsieh, C. L. The DNA methyltransferase-like protein DNMT3L stimulates de novo methylation by Dnmt3a. Proc. Natl Acad. Sci. USA 99, 16916–16921 (2002).
Bourc’his, D. & Bestor, T. H. Meiotic catastrophe and retrotransposon reactivation in male germ cells lacking Dnmt3L. Nature 431, 96–99 (2004).
Suetake, I., Shinozaki, F., Miyagawa, J., Takeshima, H. & Tajima, S. DNMT3L stimulates the DNA methylation activity of Dnmt3a and Dnmt3b through a direct interaction. J. Biol. Chem. 279, 27816–27823 (2004).
Webster, K. E. et al. Meiotic and epigenetic defects in Dnmt3L-knockout mouse spermatogenesis. Proc. Natl Acad. Sci. USA 102, 4068–4073 (2005).
Barau, J. et al. The DNA methyltransferase DNMT3C protects male germ cells from transposon activity. Science 354, 909–912 (2016).
Jain, D. et al. rahu is a mutant allele of Dnmt3c, encoding a DNA methyltransferase homolog required for meiosis and transposon repression in the mouse male germline. PLoS Genet. 13, e1006964 (2017).
Kaneda, M. et al. Essential role for de novo DNA methyltransferase Dnmt3a in paternal and maternal imprinting. Nature 429, 900–903 (2004).
Kato, Y. et al. Role of the Dnmt3 family in de novo methylation of imprinted and repetitive sequences during male germ cell development in the mouse. Hum. Mol. Genet. 16, 2272–2280 (2007).
Ozata, D. M., Gainetdinov, I., Zoch, A., O’Carroll, D. & Zamore, P. D. PIWI-interacting RNAs: small RNAs with big functions. Nat. Rev. Genet. 20, 89–108 (2019).
De Fazio, S. et al. The endonuclease activity of Mili fuels piRNA amplification that silences LINE1 elements. Nature 480, 259–263 (2011).
Vasiliauskaitė, L. et al. A MILI-independent piRNA biogenesis pathway empowers partial germline reprogramming. Nat. Struct. Mol. Biol. 24, 604–606 (2017).
Ariyoshi, M. & Schwabe, J. W. A conserved structural motif reveals the essential transcriptional repression function of Spen proteins and their role in developmental signaling. Genes Dev. 17, 1909–1920 (2003).
Mikami, S. et al. Structural insights into the recruitment of SMRT by the corepressor SHARP under phosphorylative regulation. Structure 22, 35–46 (2014).
Hata, K., Kusumi, M., Yokomine, T., Li, E. & Sasaki, H. Meiotic and epigenetic aberrations in Dnmt3L-deficient male germ cells. Mol. Reprod. Dev. 73, 116–122 (2006).
Rogakou, E. P., Pilch, D. R., Orr, A. H., Ivanova, V. S. & Bonner, W. M. DNA double-stranded breaks induce histone H2AX phosphorylation on serine 139. J. Biol. Chem. 273, 5858–5868 (1998).
Manakov, S. A. et al. MIWI2 and MILI have differential effects on piRNA biogenesis and DNA methylation. Cell Rep. 12, 1234–1243 (2015).
Watanabe, T. et al. Role for piRNAs and noncoding RNA in de novo DNA methylation of the imprinted mouse Rasgrf1 locus. Science 332, 848–852 (2011).
Aravin, A. A. et al. A piRNA pathway primed by individual transposons is linked to de novo DNA methylation in mice. Mol. Cell 31, 785–799 (2008).
Kloet, S. L. et al. Towards elucidating the stability, dynamics and architecture of the nucleosome remodeling and deacetylase complex by using quantitative interaction proteomics. FEBS J. 282, 1774–1785 (2015).
Mashtalir, N. et al. Modular organization and assembly of SWI/SNF family chromatin remodeling complexes. Cell 175, 1272–1288 (2018).
Carrieri, C. et al. A transit-amplifying population underpins the efficient regenerative capacity of the testis. J. Exp. Med. 214, 1631–1641 (2017).
Wang, H. et al. One-step generation of mice carrying mutations in multiple genes by CRISPR/Cas-mediated genome engineering. Cell 153, 910–918 (2013).
Yang, H. et al. One-step generation of mice carrying reporter and conditional alleles by CRISPR/Cas-mediated genome engineering. Cell 154, 1370–1379 (2013).
Wiśniewski, J. R., Zougman, A., Nagaraj, N. & Mann, M. Universal sample preparation method for proteome analysis. Nat. Methods 6, 359–362 (2009).
Rappsilber, J., Ishihama, Y. & Mann, M. Stop and go extraction tips for matrix-assisted laser desorption/ionization, nanoelectrospray, and LC/MS sample pretreatment in proteomics. Anal. Chem. 75, 663–670 (2003).
Hein, M. Y. et al. A human interactome in three quantitative dimensions organized by stoichiometries and abundances. Cell 163, 712–723 (2015).
Richards, A. L. et al. One-hour proteome analysis in yeast. Nat. Protoc. 10, 701–714 (2015).
Cox, J. et al. Accurate proteome-wide label-free quantification by delayed normalization and maximal peptide ratio extraction, termed MaxLFQ. Mol. Cell. Proteomics 13, 2513–2526 (2014).
The UniProt Consortium. UniProt: the universal protein knowledgebase. Nucleic Acids Res. 45 (D1), D158–D169 (2017).
Tyanova, S. et al. The Perseus computational platform for comprehensive analysis of (prote)omics data. Nat. Methods 13, 731–740 (2016).
Hubner, N. C. & Mann, M. Extracting gene function from protein-protein interactions using Quantitative BAC InteraCtomics (QUBIC). Methods 53, 453–459 (2011).
Kosugi, S., Hasebe, M., Tomita, M. & Yanagawa, H. Systematic identification of cell cycle-dependent yeast nucleocytoplasmic shuttling proteins by prediction of composite motifs. Proc. Natl Acad. Sci. USA 106, 10171–10176 (2009).
Morgan, M. et al. mRNA 3′ uridylation and poly(A) tail length sculpt the mammalian maternal transcriptome. Nature 548, 347–351 (2017).
Morgan, M. et al. A programmed wave of uridylation-primed mRNA degradation is essential for meiotic progression and mammalian spermatogenesis. Cell Res. 29, 221–232 (2019).
Sievers, F. et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 7, 539 (2011).
Kelley, L. A., Mezulis, S., Yates, C. M., Wass, M. N. & Sternberg, M. J. The Phyre2 web portal for protein modeling, prediction and analysis. Nat. Protoc. 10, 845–858 (2015).
Zhang, Y., Rataj, K., Simpson, G. G. & Tong, L. Crystal structure of the SPOC domain of the Arabidopsis flowering regulator FPA. PLoS One 11, e0160694 (2016).
Waterhouse, A. M., Procter, J. B., Martin, D. M., Clamp, M. & Barton, G. J. Jalview Version 2—a multiple sequence alignment editor and analysis workbench. Bioinformatics 25, 1189–1191 (2009).
Altschul, S. F. et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25, 3389–3402 (1997).
Aken, B. L. et al. Ensembl 2017. Nucleic Acids Res. 45 (D1), D635–D642 (2017).
Zerbino, D. R. et al. Ensembl 2018. Nucleic Acids Res. 46 (D1), D754–D761 (2018).
Karolchik, D. et al. The UCSC Table Browser data retrieval tool. Nucleic Acids Res. 32, D493–D496 (2004).
Gramates, L. S. et al. FlyBase at 25: looking to the future. Nucleic Acids Res. 45 (D1), D663–D671 (2017).
Wernersson, R. & Pedersen, A. G. RevTrans: multiple alignment of coding DNA from aligned amino acid sequences. Nucleic Acids Res. 31, 3537–3539 (2003).
Wernersson, R. Virtual Ribosome—a comprehensive DNA translation tool with support for integration of sequence feature annotation. Nucleic Acids Res. 34, W385–W388 (2006).
Goujon, M. et al. A new bioinformatics analysis tools framework at EMBL-EBI. Nucleic Acids Res. 38, W695–W699 (2010).
Ronquist, F. & Huelsenbeck, J. P. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19, 1572–1574 (2003).
Altekar, G., Dwarkadas, S., Huelsenbeck, J. P. & Ronquist, F. Parallel Metropolis coupled Markov chain Monte Carlo for Bayesian phylogenetic inference. Bioinformatics 20, 407–415 (2004).
Miller, M. A., Pfeiffer, W. & Schwartz, T. Creating the CIPRES Science Gateway for inference of large phylogenetic trees. 2010 Gateway Computing Environments Workshop (GCE) (2010).
Di Giacomo, M., Comazzetto, S., Sampath, S. C., Sampath, S. C. & O’Carroll, D. G9a co-suppresses LINE1 elements in spermatogonia. Epigenetics Chromatin 7, 24 (2014).
Kim, D., Langmead, B. & Salzberg, S. L. HISAT: a fast spliced aligner with low memory requirements. Nat. Methods 12, 357–360 (2015).
Anders, S., Pyl, P. T. & Huber, W. HTSeq—a Python framework to work with high-throughput sequencing data. Bioinformatics 31, 166–169 (2015).
Love, M. I., Huber, W. & Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014).
Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBNet Journal 17, 10–12 (2011).
Bao, W., Kojima, K. K. & Kohany, O. Repbase Update, a database of repetitive elements in eukaryotic genomes. Mob. DNA 6, 11 (2015).
Langmead, B. & Salzberg, S. L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359 (2012).
Krueger, F. & Andrews, S. R. Bismark: a flexible aligner and methylation caller for Bisulfite-Seq applications. Bioinformatics 27, 1571–1572 (2011).
Kabayama, Y. et al. Roles of MIWI, MILI and PLD6 in small RNA regulation in mouse growing oocytes. Nucleic Acids Res. 45, 5387–5398 (2017).
Langmead, B., Trapnell, C., Pop, M. & Salzberg, S. L. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10, R25 (2009).
Acknowledgements
This research was supported by Welcome Trust funding to D. O'C. (106144), R. V. B. (213612), A. G. C. (200898), J. R. (103139), R. C. A. (095021, 200885) and the Wellcome Centre for Cell Biology (203149) and a multi-user equipment grant (108504). D. O'C.’s laboratory is also supported by the European Union H2020 program grant GermAge. A. Z. was funded by a German Research Foundation fellowship (DFG, award ZO 376/1-1). We acknowledge the EMBL GeneCore facility in Heidelberg, Germany for preparing the microarray data set and RNA-seq data set of E16.5 gonocytes and sequencing all next-generation sequencing libraries, and specifically F. Jung at GeneCore for preparing the Methyl-seq libraries.
Author information
Authors and Affiliations
Contributions
A. Z. contributed to the design, execution and analysis of most experiments. T. A. helped established IP conditions and performed mass spectrometry analysis under the guidance of J. R. and R. C. A. R. B. V. and Y. K. performed bioinformatic analysis of Methyl-seq data or sRNA-seq as well as RNA-seq data, respectively. T. S. prepared sRNA-seq libraries and, with Y. K., RNA-seq libraries of P20 testes. M. H and A. G. C. performed homology alignment of the SPOC and TFIIS-M domains. L. V. performed immunofluorescence staining of HA-MIWI2 and generated the gonocyte microarray dataset. Y. A. P-R. performed phylogenetic analysis under guidance of A. S. D. O’C. conceived and supervised this study. D. O’C. and A. Z. wrote the final version of the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information Nature thanks Déborah Bourc’his, Falk Butter, Matthew Lorincz and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
Extended Data Fig. 1 Expression pattern and presence of nuclear localization signals for novel MIWI2 interactors.
a, b, Relative expression of indicated transcripts as measured by Affymetrix microarray in E16.5 gonocytes (n = 2), adult spermatogonia (n = 3), spermatocytes (n = 3), MEFs (n = 3) and bone marrow (n = 2). Data are mean and s.e.m. NLS indicates presence of a nuclear localization signal as predicted by cNLS mapper.
Extended Data Fig. 2 Homology alignment of SPOCD1 SPOC and TFIIS-M domains.
a, Multiple sequence alignment of the SPOC domain from SPOCD1 with representative vertebrate sequences from PHF3, DIDO1 and SPEN orthologues. The numbering for mouse SPOCD1 is shown above. Secondary structure elements for the human SHARP SPOC domain (PDBid 1OW1; SHARP is the human SPEN orthologue) are shown below the sequence, with dark rectangles for alpha helices and lighter arrows for beta strands. b, Multiple sequence alignment of TFIIS-M domain of SPOCD1 with equivalent sequences from TFIIS, PHF3 and DIDO1. Secondary structure elements from human PHF3 (PDBid 2DME) and human TFIIS (PDBid 3NDQ) are shown below, using the same annotation as in a. Sequences are coloured according to sequence identity.
Extended Data Fig. 3 Phylogeny of SPOCD1.
Bayesian phylogeny of Spocd1 (blue) and its vertebrate paralogs Phf3 (red) and Dido1 (green) inferred from cDNA sequences. Posterior probabilities of splits are shown as node labels. Branch lengths measure the expected substitutions per site as indicated in the scale bar.
Extended Data Fig. 4 Generation and characterization of the Spocd1null mouse allele.
a, Schematic representation of the Spocd1 locus and the encoded 1015 amino acids (aa) protein (transcript XM_017320505.1) as well as design of the sgRNA targeting Spocd1 exon 7, which harbours part of the TFIIS-M domain. b, Schematic representation and sequencing trace of the part of Spocd1− exon 7 harbouring the mutation site. The mutated site, highlighted in red, contains two premature stop codons and causes a frameshift. Sequencing was repeated with identical results on n = 3 animals. c, Representative image of genotyping results for Spocd1+/+, Spocd1+/− and Spocd1−/− animals. Similar results were obtained for all animals of the Spocd1− line. d, Numbers of E16.5 embryos per plug from matings of mice with the indicated Spocd1 genotypes. Mean and s.e.m. from n = 7 Spocd1+/+ dams mated to n = 3 Spocd1+/+ studs and n = 12 Spocd1−/− dams mated to n = 5 Spocd1+/− studs is plotted. NS, nonsignificant difference (P ≅ 0.98), two-tailed Student’s t-test. e, Representative PAS and haematoxylin stained histological testis sections of different stages of the seminiferous cycle are shown of (n = 3) Spocd1+/+ and Spocd1−/− animals, indicating a germ cell differentiation arrest at the early pachytene stage. Scale bar, 5 μm. eP, early pachytene; RS, round spermatids, eS(13), elongating spermatids (step 13); PL, pre-leptotene; P, pachytene; L, leptotene; Z, zygotene; m2, secondary meiocytes. f, Representative images of zygotene spermatocytes in wild-type and Spocd1−/− adult testis sections stained for the synaptonemal complex proteins SCP1 (red) and SCP3 (green). DNA stained with DAPI (blue). Scale bar, 1 μm. The representative images presented in panels e and f are from n = 3 mice per genotype. g, h, Analysis of transposable element expression in P20 testes from n = 3 wild-type, Spocd1−/− and Miwi2−/− mice by RNA-seq. g, Comparison of transposable element expression in Miwi2−/− and wild-type testes is shown. Transposable elements with a significantly different (P < 0.01, Benjamini–Hochberg-adjusted two-sided Wald’s test) change in expression (>2-fold) are highlighted in red and the top 12 most upregulated transposable elements in Miwi2−/− testes are labelled. h, Comparison of transposable element expression in Spocd1−/− and wild-type testes is shown. Transposable elements with a significantly different (P < 0.01, Benjamini−Hochberg-adjusted two-sided Wald’s test) change in expression (>2-fold) are highlighted in red and the same transposable elements as in a are labelled. i, Comparison of transposable element expression in Spocd1−/− and Miwi2−/− testes is shown. Transposable elements with a significantly different (P < 0.01, Benjamini−Hochberg-adjusted two-sided Wald’s test) change in expression (>2-fold) are highlighted in red. Transposable elements that are significantly upregulated in Miwi2−/− relative to wild type are highlighted in black.
Extended Data Fig. 5 CpG methylation analysis of different genomic features and transposable element families.
Analysis of genomic CpG methylation of undifferentiated P14 spermatogonia from (n = 3) wild-type, Spocd1−/− and Miwi2−/− mice is presented. a, b, Scatter plots comparing CpG methylation levels for the respective genomic features between wild-type and Spocd1−/− or Miwi2−/− (a) and between Spocd1−/− or Miwi2−/− spermatogonia (b). c, d, Scatter plots comparing CpG methylation levels for the respective transposable element families between wild-type and Spocd1−/− or Miwi2−/− (c) and between Spocd1−/− or Miwi2−/− spermatogonia (d.) Data are mean from n = 3 biological replicates per genotype and shown as individual data points (grey) overlaid by a density map.
Extended Data Fig. 6 Methylation analysis of transposable element families.
Analysis of genomic CpG methylation of undifferentiated P14 spermatogonia from (n = 3) wild-type, Spocd1−/− and Miwi2−/− mice. a, Metaplots of CpG methylation over L1Md_A, IAPEy and MMERVK10C elements and adjacent 2 kb. Below, schematic representation of the element. b, Metaplots of mean CpG methylation over LINE1 elements and adjacent 1 kb. The Methyl-seq data sets of P14 wild-type, Miwi2−/− and Spocd1−/− spermatogonia are compared to WGBS data sets of adult Mili−/− spermatocytes (Molaro et al. 20144) and P10 Dnmt3c+/−, Dnmt3c−/−, Dnmt3l+/− and Dnmt3l−/− germ cells (Barau et al. 201612). Below, schematic representation of LINE1. c, Correlation analysis of mean CpG methylation loss relative to wild type over individual elements of the indicated transposable element family in relation to their divergence from the consensus sequence for Miwi2−/− and Spocd1−/− spermatogonia.
Extended Data Fig. 7 piRNA analysis.
piRNA analyses of small RNAs sequenced from E16.5 testis from (n = 3) Spocd1+/− and Spocd1−/− mice are presented. a, Relative frequency of piRNAs mapping to LINE1 and IAP families from Spocd1+/− and Spocd1−/− E16.5 testes. Plots are shown for all piRNA or antisense piRNAs. Data are mean and s.e.m. Adjusted P-values are listed; P ≅ 1.0 values are denoted as NS (Bonferroni-adjusted two-sided Student’s t-test). b, Scatter plots showing mean expression of all (n = 124,411) piRNAs. The identity line is shown in red. r, Pearson’s correlation coefficient. c, Nucleotide features of piRNA from Spocd1+/− and Spocd1−/− E16.5 testes. Frequency of mapped piRNAs with a U at position 1 (1U) and with an A at position 10 (10A) are shown for L1Md_T elements. Data represent the mean and s.e.m. Adjusted P-values are shown (Bonferroni-adjusted two-sided Student’s t-test) d, Ping-pong analysis of piRNAs from Spocd1+/− and Spocd1−/− E16.5 testis. Relative frequencies of the distances between 5′ ends of complementary piRNAs are shown for the indicated LINE1 and IAP families. e, Nucleotide features of piRNA from Spocd1+/− and Spocd1−/− E16.5 testis. Relative frequencies of piRNAs with a U at position 1 (1U) and with an A at position 10 (10A) are shown for respective elements shown in (d). Data are mean and s.e.m. Adjusted P-values are listed; P ≅ 1.0 values are denoted as NS (Bonferroni-adjusted two-sided Student’s t-test) f, Positions of piRNAs mapped to the consensus sequence of L1Md_T. Positive and negative values indicate sense and antisense piRNAs, respectively. Schematic representation of L1Md_T is shown above.
Extended Data Fig. 8 Transposable element and gene expression in Spocd1−/− gonocytes.
Analysis of transposable element and gene expression in E16.5 Spocd1+/− and Spocd1−/− gonocytes by RNA-seq from n = 3 mice per genotype. a, Comparison of transposable element expression in Spocd1+/− and Spocd1−/− gonocytes is shown. Transposable elements upregulated in Miwi2−/− testes at P20 are highlighted in black. b, Comparison of gene expression in Spocd1+/− and Spocd1−/− gonocytes. Significantly expressed genes (P < 0.01, Benjamini–Hochberg-adjusted two-sided Wald’s test, >2-fold change) are highlighted in red.
Extended Data Fig. 9 Generation of the Spocd1HA mouse allele.
a, Schematic representation of the SPOCD1 protein and Spocd1 locus as well as design of the sgRNA targeting the 3′ UTR near the translation termination site on Spocd1 exon 15. The Spocd1HA allele encodes for a carboxy-terminal GGGGS linker followed by the HA epitope tag. The protospacer adjacent motif (PAM) site was mutated to inhibit re-targeting of the Spocd1HA allele by the sgRNA-CAS9 complex. All inserted nucleotides and corresponding encoded amino acids are highlighted in red. The SPOCD1-HA protein is shown as a schematic representation. b, Schematic representation of the targeting strategy to generate the Spocd1HA allele with a short single-stranded DNA oligo donor (ssODN) of 200 nucleotides containing 5′ and 3′ homology arms (5′HA and 3′HA) of 72 nucleotides. c, Representative image of genotyping results for Spocd1+/+, Spocd1HA/+ and Spocd1HA/HA animals. Similar results were obtained for all animals of the Spocd1HA line. d, Sequencing trace of part of a PCR amplicon of the HA epitope tag insertion site from a Spocd1HA/HA animal. The experiment was repeated with identical results on n = 2 animals. e, f, g, Representative images of wild-type (e), Spocd1HA/+ (f) and Miwi2HA/+ (g) testis sections at the indicated developmental time point probed with anti-HA antibody in green. DNA stained with DAPI in blue. Scale bars, 10 μm. The representative images presented in e–g are from experiments done in n = 3 mice as biological replicates with similar results.
Extended Data Fig. 10 Co-immunoprecipitation experiments of SPOCD1 and DNMT3A/L/C in HEK cells.
Western blot analysis of co-immunoprecipitation of SPOCD1-HA with DNMT3L-FLAG, DNMT3A-FLAG, DNMT3C-FLAG or GFP in HEK cells. Shown are lysate sample (L), control IP (protein G beads) (B) and anti-HA IP (IP) for 4 experiments. For uncropped source data, see Supplementary Fig. 1.
Supplementary information
Supplementary Information
Supplementary information containing uncropped scans of Western blot experiments shown in Extended Data Figure 10 (Supplementary Figure 1), the FACS gating strategy for sorting undifferentiated spermatogonia (Supplementary Figure 2) and seven additional data tables (Supplementary Table 1-7) relating to IPMS data, RNA-seq data, and the Methyl-seq datasets.
Rights and permissions
About this article
Cite this article
Zoch, A., Auchynnikava, T., Berrens, R.V. et al. SPOCD1 is an essential executor of piRNA-directed de novo DNA methylation. Nature 584, 635–639 (2020). https://doi.org/10.1038/s41586-020-2557-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41586-020-2557-5
This article is cited by
-
Critical appraisal of the piRNA-PIWI axis in cancer and cancer stem cells
Biomarker Research (2024)
-
Small RNAs, spermatogenesis, and male infertility: a decade of retrospect
Reproductive Biology and Endocrinology (2023)
-
Transcriptomic responses in the blood and sputum of cigarette smokers compared to e-cigarette vapers
Respiratory Research (2023)
-
The emerging role of the piRNA/PIWI complex in respiratory tract diseases
Respiratory Research (2023)
-
The epigenetic regulatory mechanism of PIWI/piRNAs in human cancers
Molecular Cancer (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.