Abstract
Cancer cachexia is a highly prevalent condition associated with poor quality of life and reduced survival1. Tumor-induced perturbations in the endocrine, immune and nervous systems drive anorexia and catabolic changes in adipose tissue and skeletal muscle, hallmarks of cancer cachexia2,3,4. However, the molecular mechanisms driving cachexia remain poorly defined, and there are currently no approved drugs for the condition. Elevation in circulating growth differentiation factor 15 (GDF15) correlates with cachexia and reduced survival in patients with cancer5,6,7,8, and a GDNF family receptor alpha like (GFRAL)–Ret proto-oncogene (RET) signaling complex in brainstem neurons that mediates GDF15-induced weight loss in mice has recently been described9,10,11,12. Here we report a therapeutic antagonistic monoclonal antibody, 3P10, that targets GFRAL and inhibits RET signaling by preventing the GDF15-driven interaction of RET with GFRAL on the cell surface. Treatment with 3P10 reverses excessive lipid oxidation in tumor-bearing mice and prevents cancer cachexia, even under calorie-restricted conditions. Mechanistically, activation of the GFRAL–RET pathway induces expression of genes involved in lipid metabolism in adipose tissues, and both peripheral chemical sympathectomy and loss of adipose triglyceride lipase protect mice from GDF15-induced weight loss. These data uncover a peripheral sympathetic axis by which GDF15 elicits a lipolytic response in adipose tissue independently of anorexia, leading to reduced adipose and muscle mass and function in tumor-bearing mice.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data Availability
The co-ordinates of the GFRAL–3P10 crystal structure are available in the Protein Data Bank (http://www.rcsb.org/pdb), with PDB accession codes 6WMW. Data collection, phasing and refinement statistics are included in Supplementary Table 4. RNA-sequencing data are available on Gene Expression Omnibus (GEO), with accession number GSE149263.
References
Tisdale, M. J. Molecular pathways leading to cancer cachexia. Physiol. (Bethesda) 20, 340–8 (2005).
Baracos, V. E., Martin, L., Korc, M., Guttridge, D. C. & Fearon, K. C. H. Cancer-associated cachexia. Nat. Rev. Dis. Prim. 4, 17105 (2018).
Burfeind, K. G., Michaelis, K. A. & Marks, D. L. The central role of hypothalamic inflammation in the acute illness response and cachexia. Semin Cell Dev. Biol. 54, 42–52 (2016).
Argiles, J. M., Busquets, S., Stemmier, B. & Lopez-Soriano, F. J. Cancer cachexia: understanding the molecular basis. Nat. Rev. Cancer 14, 754–762 (2014).
Johnen, H. et al. Tumor-induced anorexia and weight loss are mediated by the TGF-β superfamily cytokine MIC-1. Nat. Med. 13, 1333–1340 (2007).
Lerner, L. et al. Plasma growth differentiation factor 15 is associated with weight loss and mortality in cancer patients. J. Cachexia Sarcopenia Muscle 6, 317–324 (2015).
Staff, A. C. et al. Growth differentiation factor-15 as a prognostic biomarker in ovarian cancer. Gynecol. Oncol. 118, 237–43 (2010).
Li, C. et al. Growth differentiation factor 15 is a promising diagnostic and prognostic biomarker in colorectal cancer. J. Cell Mol. Med. 20, 1420–6 (2016).
Hsu, J. et al. Non-homeostatic body weight regulation through a brainstem-restricted receptor for GDF15. Nature 550, 255–259 (2017).
Mullican, S. et al. GFRAL is the receptor for GDF15 and the ligand promotes weight loss in mice and nonhuman primates. Nat. Med. 23, 1150–1157 (2017).
Yang, L. et al. GFRAL is the receptor for GDF15 and is required for the anti-obesity effects of the ligand. Nat. Med. 23, 1158–1166 (2017).
Emmerson, P. J. et al. The metabolic effects of GDF15 are mediated by the orphan receptor GFRAL. Nat. Med. 23, 1215–1219 (2017).
Treanor, J. J. et al. Characterization of a multicomponent receptor for GDNF. Nature 382, 80–3 (1996).
Klein, R. D. et al. A GPI-linked protein that interacts with Ret to form a candidate neurturin receptor. Nature 387, 717–21 (1997).
Lie, J. et al. Cryo-EM analyses reveal the common mechanism and diversification in the activation of RET by different ligands. Elife 19, e47650 (2019).
Parkash, V. et al. The structure of the glial cell line-derived neurotrophic factor-coreceptor complex: insights into RET signaling and heparin binding. J. Biol. Chem. 283, 35164–72 (2008).
De Vita, G. et al. Tyrosine 1062 of RET–MEN2A mediates activation of Akt (protein kinase B) and mitogen-activated protein kinase pathways leading to PC12 cell survival. Cancer Res. 60, 3727–3731 (2000).
Lerner, L. et al. MAP3K11/GDF15 axis is a critical driver of cancer cachexia. J. Cachexia Sarcopenia Muscle 7, 467–82 (2016).
Coll, A. et al. GDF15 mediates the effects of metformin on body weight and energy balance. Nature 578, 444–448 (2020).
Das, S. K. et al. Adipose triglyceride lipase contributes to cancer-associated cachexia. Science 333, 233–8 (2011).
Agustsson, T. et al. Mechanism of increased lipolysis in cancer cachexia. Cancer Res. 67, 5531–7 (2007).
Rydén, M. et al. Lipolysis—not inflammation, cell death, or lipogenesis—is involved in adipose tissue loss in cancer cachexia. Cancer 113, 1695–704 (2008).
Evans, W. J. et al. Cachexia: a new definition. Clin. Nutr. 27, 793–799 (2008).
Tsai, V. W. W., Husaini, Y., Sainsbury, A., Brown, D. A. & Breit, S. N. The MIC-1/GDF15–GFRAL pathway in energy homeostasis: implications for obesity, cachexia, and other associated diseases. Cell Metab. 28, 353 (2018).
Campos, C. A., Bowen, A. J., Roman, C. W. & Palmiter, R. D. Encoding of danger by parabrachial CGRP neurons. Nature 555, 617–622 (2018).
Luan, H. H. et al. GDF15 is an inflammation-induced central mediator of tissue tolerance. Cell 178, 1231–1244 (2019).
Fukawa, T. et al. Excessive fatty acid oxidation induces muscle atrophy in cancer cachexia. Nat. Med. 22, 666–671 (2016).
Holm, L. & Kjaer, M. Measuring protein breakdown in individual proteins in vivo. Curr. Opin. Clin. Nutr. Metab. Care. 13, 526–531 (2010).
Strelau, J. et al. Progressive postnatal motoneuron loss in mice lacking GDF-15. J. Neurosci. 29, 13640–13648 (2009).
Zeisel, A. et al. Molecular architecture of the mouse nervous system. Cell 174, 999–1014 (2018).
Robinson, M. D., McCarthy, D. J. & Smyth, G. K. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140 (2010).
McCarthy, D. J., Chen, Y. & Smyth, G. K. Differential expression analysis of multifactor RNA-seq experiments with respect to biological variation. Nucleic Acids Res. 40, 4288–4297 (2012).
Acknowledgements
We thank J. Lichtman for help with statistical analyses of data, L. Cheng for help with in vitro complex-formation assays and E. Estrin for the cloning of various GFRAL point mutants.
Author information
Authors and Affiliations
Contributions
R.S., J.T., D.A.L. and B.B.A. directed the work. R.S., D.A.L. and B.B.A. designed experiments, analyzed data and wrote the manuscript. Y.W. and B.L. generated antibodies. J.O., R.V., V.P., D.A., J.-Y.H. and S.R.S. developed and performed HTRF, ELISAs, NanoBiT, cell-based signaling and reporter and FACS assays. M.M. and M.J.S. designed, performed and quantified immunofluorescence experiments in brain sections. M.C. designed, managed and performed mouse experiments along with Z.G., P.T and S.D.K. M.W. and A.K. created all expression constructs. D.L., J.H. and R.H. expressed and purified recombinant proteins and antibodies. J.H. generated crystals and performed in vitro complex formation assays. Surface plasmon resonance experiments were performed by K.M. Crystal structures were generated and solved by D.L., A.O. and A.W. RNA-sequencing analyses were performed by H.-I H.C.
Corresponding author
Ethics declarations
Competing interests
D.L., A.W. and A.O. declare no direct competing financial interests. All other authors are, or were, employees of NGM Biopharmaceuticals and may hold stock or stock options in this company. NGM Biopharmaceuticals, Inc. has obtained a patent, entitled “Binding proteins and methods of use thereof” (US10174119B2), that discloses protein sequences of the anti-GFRAL antibodies, GFRAL–antibody protein complexes and potential uses of those agents. J.T. and Y.W. are listed as inventors.
Additional information
Peer review information Javier Carmona was the primary editor on this article, and managed its editorial process and peer review in collaboration with the rest of the editorial team
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 3P10 does not bind GFRα1 and does not inhibit GDNF/GFRα1 or Neurturin/GFRα2 signaling.
a, Sensorgrams of GFRAL (0.31-20 nM) binding to 3P10 (left); steady state affinity curve fit (right). RU, response unit. Data representative of three experimental replicates. b, Sensorgrams of 3P10 binding GFRAL (left) and not GFRa1 (middle). GFRa1 binds GDNF (right). Data representative of two experimental replicates. c, Western blot for total and phosphorylated ERK (pERK) from GFRAL and RET transfected 293 T cells incubated with GDNF or Neurturin and 3P10. Representative gel from three independent experiments.
Extended Data Fig. 2 3P10 binds GFRAL at its interface with RET and GFRAL Arg294 is required for 3P10 binding.
a, A 2.97 Å crystal structure of 3P10 in complex with GFRAL ECD, with GDF15 docked from 5VZ4.pdb. b, The GFRAL ECD interface with 3P10 shares 5 common residues (Arg294, Thr295, Thr297, Gln298, Ser299) with the GFRAL ECD interface with RET ECD from 6Q2J.pdb, based on structural docking. c, 3P10 HC-Tyr100 and 3P10 LC-Tyr53 in an edge-to-face pi-pi stacking interaction with GFRAL Arg294 (yellow). d, FACS plots of 3P10 binding to cells expressing wildtype GFRAL but not to cells expressing Arg294Glu mutant GFRAL. 25M22-FITC binding to cells expressing both wildtype GFRAL and the Arg294Glu mutant GFRAL are shown as a control for cell surface expression. Data representative of two independent experiments. e, GDF15 reporter gene assay of cells expressing wildtype or Arg294Glu mutant GFRAL incubated with increasing concentrations of 3P10. N = 3 per condition. Data represent mean ± s.d.
Extended Data Fig. 3 3P10 reverses tumor-induced tissue and body weight loss in GDF15 expressing Patient Derived Xenograft (PDX) and syngeneic mouse models.
a-c, Quantification of serum GDF15 from HT1080 (a), syngeneic (b) and patient derived xenograft (c) tumor-bearing mice. Data represent mean ± s.e.m. In (a) data represent n = 6 mice/group. In (c) data represent n = 10 for non-tumor bearing (NTB), n = 6 for gastric, n = 9 for ovarian and n = 12 for liver PDX models. d,e Tumor growth measured by caliper of KLN205 and Ovarian PDX shown in Fig. 2e,f. n = 10 per group. f-i, Change in body weight and tumor volume of syngeneic RENCA (f) and B16Bl6 models (g), and gastric (h) and liver (i) PDX models treated with IgG (black) or 3P10 (blue). Data represent mean ± s.e.m. Data represent n = 10 mice/group for (f,g), n = 20 mice/group for (h) and n = 12 mice/group for (i). Numbers above indicate p-values calculated by two-sided t-test. Body weight (BW) was calculated by subtracting the estimated tumor weight (1 g = 1000mm3) from the total, measured body weight to account for variability in tumor size. j, Tissue weights measured from gastric and ovarian PDX tumor models at end of study. Center values indicate mean. Data represent n = 20/group for gastric and n = 9 for ovarian PDX models. Numbers above indicate p-values calculated by two-sided t-test. k, Body weight of normal chow mice (n = 8 per group) treated weekly with IgG1 or 3P10 antibody. Data represent mean +/− s.e.m. All antibodies were dosed at 3 mg/kg in tumor-bearing mice at the times depicted by the arrowheads.
Extended Data Fig. 4 3P10 reverses HT1080 tumor-induced tissue and body weight loss.
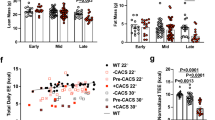
a,b, Cumulative food intake after 1 mg/kg recombinant GDF15 (a) and inoculation with HT1080 tumor cells (b). n = 10 mice per group. Data represent mean ± SEM. Numbers above indicate p-values calculated by two-sided t-test. c, Body weights of non-tumor bearing (NTB) and HT1080 mice treated on day 11 with 3 mg/kg antibody, fed ad libidum without calorie restriction. Data represent mean ± SEM. n = 10 mice per group. Number above indicate p-values calculated by two-sided t-test. d, White adipose and liver weight on day 21 (refer to c). n = 5 per group. Data represent mean ± s.e.m. Numbers above denote p-values calculated by ANOVA. e, 24 hour food intake measured 1 or 2 days after antibody treatment (days 11-13 in c). Data represent mean ± s.e.m. On day 1, n = 8 for non-tumor bearing (NTB) and n = 9 for IgG1 and 3P10. On day 2, n = 5 for NTB and n = 4 for IgG1 and 3P10. f, Fat and lean mass measured on day 21 by MRI of calorie restricted, IgG1 and 3P10-treated HT1080 animals (n = 10) after tumors removed (See Fig. 3b). Numbers above indicates p-values calculated by two-sided t-test. Data represent mean ± s.e.m.
Extended Data Fig. 5 Chemical peripheral sympathectomy with 6OHDA does not impact GLP-1-driven food intake reduction and body weight loss.
Cumulative 6 day food take a, and percent change in body weight from baseline b, of 6-OHDA treated mice after a single intraperitoneal dose of 1 mg/kg GLP-1. n = 7 mice per group. Data represent mean ± SEM. Numbers above denote p-values calculated by ANOVA.
Extended Data Fig. 6 3P10 blocks tumor-induced muscle wasting and preserves muscle function.
a, Gastrocnemius (top), tibialis anterior (middle) and soleus (bottom) muscle mass measured on day 22 from non-tumor bearing (NTB) and HT1080 tumor-bearing mice treated with 3 mg/kg IgG1 or 3P10 (See Extended Data Fig. 4c). n = 10 per group. b, Serum 3-methyl-histidine, a biomarker for muscle breakdown, 24 hours after antibody treatment of HT1080 tumor-bearing mice treated with 3 mg/kg IgG1 or 3P10, and non-tumor bearing controls. n = 6 per group. c, Differentially expressed genes identified by RNA sequencing of gastrocnemius muscle of IgG1 treated HT1080 tumor-bearing compared to non-tumor bearing mice, that are reversed by 3P10. n = 5 per group. Muscle was collected on day 22 (See Extended Data Fig. 4c). Muscle specific genes identified by Ingenuity Pathway Analysis are listed, including markers for muscle atrophy (bold). See Extended Table 3 for full list. d, Fbxo32 (top) and TRIM63 (bottom) muscle atrophy gene expression measured by RT-qPCR of HT1080 tumor bearing mice treated with 3 mg/kg IgG1 or 3P10 antibody. n = 6 per group. Gene expression was assessed on day 22 (See Extended Data Fig. 4c). e, Muscle function measured by grip strength measured on day 22 from non-tumor bearing (NTB) and HT1080 tumor-bearing mice treated with 3 mg/kg IgG1 or 3P10 (See Extended Data Fig. 4c). Bar graph represents n = 150 measurements collected from 6 mice, each of which 25 repeat measures of grip strength were counted. In a,b and d,e, data represent mean ± s.e.m. Numbers above denote p-values analyzed by ANOVA.
Supplementary information
Supplementary Information
Supplementary Tables 1–4.
Rights and permissions
About this article
Cite this article
Suriben, R., Chen, M., Higbee, J. et al. Antibody-mediated inhibition of GDF15–GFRAL activity reverses cancer cachexia in mice. Nat Med 26, 1264–1270 (2020). https://doi.org/10.1038/s41591-020-0945-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41591-020-0945-x