Abstract
Advances in genetics and sequencing have identified a plethora of disease-associated and disease-causing genetic alterations. To determine causality between genetics and disease, accurate models for molecular dissection are required; however, the rapid expansion of transcriptional populations identified through single-cell analyses presents a major challenge for accurate comparisons between mutant and wild-type cells. Here we generate mouse models of human severe congenital neutropenia (SCN) using patient-derived mutations in the GFI1 transcription factor. To determine the effects of SCN mutations, we generated single-cell references for granulopoietic genomic states with linked epitopes1, aligned mutant cells to their wild-type equivalents and identified differentially expressed genes and epigenetic loci. We find that GFI1-target genes are altered sequentially, as cells go through successive states of differentiation. These insights facilitated the genetic rescue of granulocytic specification but not post-commitment defects in innate immune effector function, and underscore the importance of evaluating the effects of mutations and therapy within each relevant cell state.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
scRNA-seq, CITE-seq, scATAC-seq and ChIP–seq data are deposited as Gene Expression Omnibus (GEO) SuperSeries GSE120409 and in Synapse (https://www.synapse.org/#!Synapse:syn16806696). Proteomic data are available via ProteomeXchange (www.ebi.ac.uk/pride/archive/) with identifier PXD010943. Extensive processed and primary data are provided and organized in Synapse, including 10x Genomics count matrices, Fluidigm expression, single-cell populations, quality control metrics, 10x Genomics Cell Ranger outputs (summary report and loupe browser files), genomic coordinate peak files and differential expression results (cellHarmony).
Code availability
All described code is provided in Github (https://github.com/nsalomonis/altanalyze).
References
Stoeckius, M. et al. Simultaneous epitope and transcriptome measurement in single cells. Nat. Methods 14, 865–868 (2017).
Gilbert, S. F. Developmental Biology, 6th edition (Sinauer Associates, 2000).
DePasquale, E. A. K. et al. cellHarmony: cell-level matching and holistic comparison of single-cell transcriptomes. Nucleic Acids Res. 47, e138 (2019).
Stuart, T. et al. Comprehensive integration of single-cell data. Cell 177, 1888–1902.e1821 (2019).
Xia, J. et al. Prevalence of mutations in ELANE, GFI1, HAX1, SBDS, WAS and G6PC3 in patients with severe congenital neutropenia. Br. J. Haematol. 147, 535–542 (2009).
Person, R. E. et al. Mutations in proto-oncogene GFI1 cause human neutropenia and target ELA2. Nat. Genet. 34, 308–312 (2003).
Wang, H. et al. A reporter mouse reveals lineage-specific and heterogeneous expression of IRF8 during lymphoid and myeloid cell differentiation. J. Immunol. 193, 1766–1777 (2014).
Deshmukh, H. S. et al. The microbiota regulates neutrophil homeostasis and host resistance to Escherichia coli K1 sepsis in neonatal mice. Nat. Med. 20, 524–530 (2014).
Luedi, P. P. et al. Computational and experimental identification of novel human imprinted genes. Genome Res. 17, 1723–1730 (2007).
Bonthuis, P. J. et al. Noncanonical genomic imprinting effects in offspring. Cell Rep. 12, 979–991 (2015).
Hock, H. et al. Intrinsic requirement for zinc finger transcription factor Gfi-1 in neutrophil differentiation. Immunity 18, 109–120 (2003).
Belyaev, N. N. et al. Induction of an IL7-R+c-Kithi myelolymphoid progenitor critically dependent on IFN-gamma signaling during acute malaria. Nat. Immunol. 11, 477–485 (2010).
MacNamara, K. C. et al. Infection-induced myelopoiesis during intracellular bacterial infection is critically dependent upon IFN-γ signaling. J. Immunol. 186, 1032–1043 (2011).
Mempel, K., Pietsch, T., Menzel, T., Zeidler, C. & Welte, K. Increased serum levels of granulocyte colony-stimulating factor in patients with severe congenital neutropenia. Blood 77, 1919–1922 (1991).
Hock, H. et al. Gfi-1 restricts proliferation and preserves functional integrity of haematopoietic stem cells. Nature 431, 1002–1007 (2004).
Karsunky, H. et al. Inflammatory reactions and severe neutropenia in mice lacking the transcriptional repressor Gfi1. Nat. Genet. 30, 295–300 (2002).
Zeng, H., Yücel, R., Kosan, C., Klein-Hitpass, L. & Möröy, T. Transcription factor Gfi1 regulates self-renewal and engraftment of hematopoietic stem cells. EMBO J. 23, 4116–4125 (2004).
Abe, T. et al. Visualization of cell cycle in mouse embryos with Fucci2 reporter directed by Rosa26 promoter. Development 140, 237–246 (2013).
Olsson, A. et al. Single-cell analysis of mixed-lineage states leading to a binary cell fate choice. Nature 537, 698–702 (2016).
Yáñez, A. et al. Granulocyte-monocyte progenitors and monocyte-dendritic cell progenitors independently produce functionally distinct monocytes. Immunity 47, 890–902.e4 (2017).
Ng, L. G., Ostuni, R. & Hidalgo, A. Heterogeneity of neutrophils. Nat. Rev. Immunol. 19, 255–265 (2019).
Farrell, J. A. et al. Single-cell reconstruction of developmental trajectories during zebrafish embryogenesis. Science 360, eaar3131 (2018).
Zhu, Y. P. et al. Identification of an early unipotent neutrophil progenitor with pro-tumoral activity in mouse and human bone marrow. Cell Rep. 24, 2329–2341.e8 (2018).
Drissen, R. et al. Distinct myeloid progenitor-differentiation pathways identified through single-cell RNA sequencing. Nat. Immunol. 17, 666–676 (2016).
Kim, M. H. et al. A late-lineage murine neutrophil precursor population exhibits dynamic changes during demand-adapted granulopoiesis. Sci. Rep. 7, 39804 (2017).
Evrard, M. et al. Developmental analysis of bone marrow neutrophils reveals populations specialized in expansion, trafficking, and effector functions. Immunity 48, 364–379.e8 (2018).
Venkatasubramanian, M., Chetal, K., Schnell D., Atluri, G. & Salomonis, N. Resolving single-cell heterogeneity from hundreds of thousands of cells through sequential hybrid clustering and NMF. Bioinformatics btaa201 (2020).
Fang, R. et al. Fast and accurate clustering of single cell epigenomes reveals cis-regulatory elements in rare cell Types. Preprint at https://www.bioRxiv.org/content/10.1101/615179v2 (2019).
Zweidler-Mckay, P. A., Grimes, H. L., Flubacher, M. M. & Tsichlis, P. N. Gfi-1 encodes a nuclear zinc finger protein that binds DNA and functions as a transcriptional repressor. Mol. Cell. Biol. 16, 4024–4034 (1996).
Gillet, L. C. et al. Targeted data extraction of the MS/MS spectra generated by data-independent acquisition: a new concept for consistent and accurate proteome analysis. Mol. Cell Proteomics 11, O111.016717 (2012).
Elsner, J., Roesler, J., Emmendörffer, A., Lohmann-Matthes, M. L. & Welte, K. Abnormal regulation in the signal transduction in neutrophils from patients with severe congenital neutropenia: relation of impaired mobilization of cytosolic free calcium to altered chemotaxis, superoxide anion generation and F-actin content. Exp. Hematol. 21, 38–46 (1993).
Holtschke, T. et al. Immunodeficiency and chronic myelogenous leukemia-like syndrome in mice with a targeted mutation of the ICSBP gene. Cell 87, 307–317 (1996).
Meyer, S. E. et al. DNMT3A haploinsufficiency transforms FLT3ITD myeloproliferative disease into a rapid, spontaneous, and fully penetrant acute myeloid leukemia. Cancer Discov. 6, 501–515 (2016).
Nayak, R. C. et al. Pathogenesis of ELANE-mutant severe neutropenia revealed by induced pluripotent stem cells. J. Clin. Invest. 125, 3103–3116 (2015).
Acknowledgements
We thank H. Singh for GFI1 antisera, H. C. Morse for supplying Irf8-GFP mice, members of the Cincinnati Children’s Hospital Medical Center (CCHMC) DNA sequencing and Genotyping Core and the Research Flow Cytometry Core (supported in part by National Institutes of Health (NIH) grants AR-47363, NIH DK78392 and NIH DK90971), and A. Sproles for assistance with cytokine analyses; S. Smith, H. Chi Liang and K. Rangel in the CCHMC Gene Expression Core for generating scRNA-seq libraries; T. Newkold for technical assistance; and H. Singh, J. Cancelas, S. Kogan, R. Kopan, J. Wells and T. Gruber for questions and discussions. This work was partly funded by support from NIH DP1AI131080 (S.S.W.) T32 ES007250 (to D.E.M.), S10RR027015 (to K.D.G.), and R01HL122661 (H.L.G.).
Author information
Authors and Affiliations
Contributions
D.E.M., G.P., S.S.W. and H.L.G. designed experiments. D.E.M., A.O., G.P., R.S., S.C. and P.D. performed experiments and also analysed data with K.G. K.Z., J.Y. and K.C.M. provided patient samples and data. L.R.T. and C.L. generated and cultured iPS cells. J.M. and D.J.K. generated the Gfi1N382S, Gfi1K403R and Gfi1R412X mice. N.S. conceived and developed the cellHarmony software with considerable input from H.L.G. S.H. and N.S. developed the gene viewer. K.F., B.S., K.C., K.L.N., N.S. and H.L.G. analysed bioinformatics data. D.E.M., N.S. and H.L.G. wrote the paper.
Corresponding authors
Ethics declarations
Competing interests
D.E.M. is currently employed by Eli Lilly and Company and K.L.N. is employed by BioLegend Inc. The remaining authors declare no competing interests.
Additional information
Peer review information Nature thanks Klaus Ley, Cornelis Murre and Leonard Zon for their contribution to the peer review of this work.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
Extended Data Fig. 1 Functional assessment of GFI1 variants derived from patients with SCN and generation of GFI1 ZnF-mutant mice.
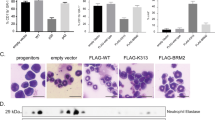
a, Representative FACS plots of lentiviral-transduced LSK cells isolated from adult Irf8-eGFP transgenic mice with the percentage of IRF8–eGFPhigh cells indicated. b, Graphical summary of FACS analysis of lentiviral-transduced LSK cells isolated from adult Irf8-eGFP transgenic mice (top) with locations of the variants mapped to the GFI1 protein (bottom). ATG, start codon; EV, empty vector; TGA, stop codon; *P2A, mutation not found in patients; SNAG; SNAIL/GFI1 family domain; SUMO, sumoylation domain. Italic font denotes other variants detected in the same patient; bold font denotes variants also found in patients diagnosed with a malignancy; grey blocks denote characterized protein domains. c, Schematic of the Gfi1 locus annotated with relevant features. Line with small arrows denote intronic regions; numbered blocks denote exons; black blocks denote coding regions; grey blocks denote noncoding regions; large arrows denote noncoding region used for genotyping. ZFN, zinc-finger nucleases. d–f, Schematic of the nucleotide changes made to the coding region and 3′ untranslated region (UTR) to introduce the N382S (d, d’), K403R (e, e’), and R412X (f, f’) mutations. g, Representative genotyping of the ZnF-mutant mice with or without restriction enzyme digestion. h, Representation of Sanger sequencing analysis of cDNA from adult whole bone marrow (WBM). Targeted wild-type nucleotides are underlined; mutated nucleotides are in bold; and red nucleotides indicate the location of the stop codon. i, Immunoblot analysis of two R412X founder lines using bone marrow lysates from adult mouse Linneg bone marrow. j, Graphical summary of FACS analysis of peripheral blood from neonatal mice. k, Total cell counts per millilitre of neonatal peripheral blood as determined by FACS. l, Representative RNAscope images (left) and transcript quantification (right) of the indicated transcripts in primary human CD34+ cells. The number of cells scored (left) and the number of donors tested (right) are indicated. m, Representative elecropherogram plots from Sanger sequencing of GFI1 RT–PCR products derived from iPS cells. The arrow indicates the nucleotide substitution made to introduce the R412X mutation. n, Representative FACS plots of iPS cells at the end of the 10-day haematopoietic differentiation protocol. o, Representative cytospin images of neutrophils derived from iPS cells. Data in a, b and l are representative of three biological replicates; data in j and k are representative of individual biological replicates; data in i display two biological replicates for each founder line; data in m–o are representative of two independent experiments. Data are mean (a) or mean ± s.e.m. (b, j–l). *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001, two-tailed t-test. Scale bars, 2 μm (l) and 10 μm (o).
Extended Data Fig. 2 Characterization of GFI1 ZnF-mutant mice at the steady-state.
a, Graphical representation of FACS analysis of peripheral blood from adult mice. b, FACS plots and graphical representation of FACS analysis of peripheral blood from adult mice. c, Graphical summary of CFU assays performed on adult mouse Linneg bone marrow from two different founder lines. Total cell counts of whole bone marrow obtained from two femurs and tibias per adult mouse. e, FACS plots and quantification of bone marrow populations from adult mice. f, Representative cytospins of whole bone marrow from adult mice. Data in f are representative of three biological replicates. Data are mean ± s.e.m. (a–e). *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001, two-tailed t-test. Scale bar, 10 μm.
Extended Data Fig. 3 Inflammatory-cytokine-independent emergency granulopoiesis and high G-CSF levels in GFI1 ZnF-mutant mice at steady-state and functional analysis of GFI1 ZnF mutants.
a, FACS plots and quantification of bone marrow cells from adult Linneg, KIT+, SCA-1+ mice. b, Quantification of adult peripheral blood cytokine levels at steady-state by cytokine array. c, Quantification of individual adult peripheral blood cytokine levels at steady-state by Luminex analysis. d, Survival analysis of lethally irradiated BoyJ recipients of adult whole bone marrow from adult GFI1-mutant donors. e, Graphical representation of FACS analysis of peripheral blood chimerism in transplant recipients from d four months after transplant. f, g, Graphical representations of the total number of colonies in mouse organs obtained 24 h after infection with an LD50 dose of C. albicans (f) or 5 × 107 CFU of S. aureus (g). h, Immunoblot analysis of adult mouse Linneg bone marrow. Data are mean ± s.e.m. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001, two-tailed t-test (a–c, e–g) or two-sided Mantel–Cox test (d). Data in h are representative of three independent experiments.
Extended Data Fig. 4 FACS and Fluidigm-based analysis of steady-state terminal granulopoiesis.
a, FACS plots demonstrating the gating strategy used for sorting. The populations sorted for scRNA-seq are indicated in shades of grey with accompanying cytospins. b, Violin plots of the gene and read-level metrics for each of the indicated libraries. Dashed lines indicate mean, lower and upper quartiles. Sample size (n = number of cells) is displayed at the top. c, Heat map of gene expression defined by ICGS (Fluidigm C1, excluding cell-cycle genes) in scRNA-seq data (n = 516 cells). Each column represents a single cell and each row represents a single gene. ICGS clusters are annotated (top). NK, natural killer T-cell progenitor. d, Joint UMAP plot of scRNA-seq data from c, separated as previously described19 and new Fluidigm captures show no detectable batch effects. e, Bar chart of the heat map in c displaying the incidence and amplitude of selected genes. f, Heat map of correlation between gene expression and each displayed cluster as generated by MarkerFinder (AltAnalyze software). g, FACS plots comparing expression of LY6G with the indicated surface marker. FMO, fluorescence minus one control. h, Heat map of cell cycle gene expression in ICGS-defined clusters in scRNA-seq data (n = 509 cells). Each column represents a single cell and each row represents a single gene. Gene expression clusters were generated in AltAnalyze and the ICGS clusters are annotated (top). FACS gates are annotated (bottom). LK CD34+, Linneg KIT+ SCA-1neg CD34+. Key genes are indicated (right). i, Scatter plot representation of scRNA-seq data from h comparing the gene expression of G1-to-S phase transition genes with G2-to-M phase transition genes in each cell. Each point represents a single cell. j, Bar chart of the heat map in h, displaying the incidence and amplitude of selected genes. k, Heat map of correlation between gene expression and each displayed cluster as generated by MarkerFinder. l, Heat map of enrichment for Gene Ontology biological processes enriched in the granulocytic clusters from the Fluidigm scRNA-seq data from c with key processes indicated (right). m, Scatter plot representation of scRNA-seq data from c, in which each point represents a single cell. Reads per cell indicate RNA-seq by expectation–maximization (RSEM) transcript-aligned read counts for each cell library. Genes expressed per cell indicate the number of genes with a transcripts per million (TPM) >1 for each single-cell library. Data in a and g are representative of three biological replicates; data in f and k display Pearson correlation values.
Extended Data Fig. 5 CITE-seq analysis of steady-state terminal granulopoiesis.
a, FACS plots demonstrating the gating strategy used for sorting. The population sorted for scRNA-seq is indicated in red. b, Heat map of gene expression for cellHarmony-assigned cell populations from CITE-seq 10x Genomics captures (male and female mice, n = 11,132 cells) compared with ICGS-defined clusters from the Fluidigm scRNA-seq data. De novo marker genes (MarkerFinder) for each assigned cluster from the 10x Genomics data are shown (top). Each column represents a single cell and each row represents a single gene. Multi-Lin*, multi-lineage primed. The gender of the host mouse of each cellular barcode (bottom) and example MarkerFinder genes in common between Fluidigm and 10x Genomics data are indicated (right). c, Violin plots of the gene and read-level metrics for each of the indicated libraries. Dashed lines indicate mean, lower, and upper quartiles. Sample size (n = number of cells) displayed (top). d, Lineage-priming scores for monocytic and granulocytic specification. Scatter plot displaying assigned scores for cellHarmony-assigned neutrophil progenitors (proNeu-2), monocytic progenitors (Mono), bi-potential monocytic–granuloctyic intermediates (IG2) and megakaryocyte progenitors (Meg) from b (see Methods). Each point represents a single cell. e, Heat map of row-normalized ADT UMI counts (log2-transformed, median subtracted) for each corresponding cell from b. f, Heat map of cell cycle gene expression displaying the same cells as b and the same genes as Extended Data Fig. 4h. g, Scatter plot representation of scRNA-seq data from f comparing the gene expression of G1-to-S phase transition genes with G2-to-M phase transition genes in each cell. Each point represents a single cell. h, Bar chart of the heat map in b, displaying the incidence and amplitude of selected genes. i, Heat map of correlation between gene expression and each displayed cluster as generated by the MarkerFinder feature of AltAnalyze. j, k, Plots of CITE-seq ADT UMI counts, in which grey indicates all captured cells, and red indicates either cells classified by ICGS clusters (j) or the top 1% of UMIs expressing the indicated gene (k). l, UMAP of 15,968 published cell-barcodes23 coloured according to cellHarmony assigned cell populations (10x Genomics reference) where each dot represents a single cell. Minor contaminant populations (<30 cells per cluster) were excluded. m, Heat map of row normalized CLR-transformed ADT counts of the indicated cell surface proteins (left) displayed as an average of all cells in the indicated clusters (top). n, Correlation plots between ADT UMI counts and the expression of genes (cellular barcode normalized UMI counts) encoding the corresponding proteins detected via CITE-seq. Each dot represents a single cell. Linear trend lines for all cells with RNA expression > 0 are indicated by dotted red lines with corresponding coefficients of determination displayed. Data in i display Pearson correlation values.
Extended Data Fig. 6 Fluidigm-based transcriptional analysis of Gfi1R412X/− cells.
a, FACS plots of adult mouse bone marrow (left) and representative cytospin of FACS-sorted adult mouse bone marrow (right). The populations sorted for scRNA-seq are indicated in red. b, Schematic summary of the cellHarmony algorithm. c, cellHarmony-assigned ICGS states of combined wild-type and Gfi1R412X/− Fluidigm scRNA-seq data (n = 624 cells, excluding cell-cycle genes). Each tick mark represents data from a single cell. Gene expression clusters were generated in AltAnalyze and the ICGS clusters are annotated (top) (Extended Data Fig. 4c). FACS gates are annotated (right). d, Population distribution of Fluidigm scRNA-seq data from c and Extended Data Fig. 4c. e, cellHarmony heat map of wild-type-aligned Gfi1R412X/− cell gene expression as ordered in c (dotted lines). Each column represents a single cell and each row represents a single gene from Extended Data Fig. 4c. The FACS gates and ICGS clusters are annotated (top) and gene clusters are indicated (left). f, Bar chart of the heat map in e, displaying the incidence and amplitude for selected genes. Arrows indicate differential expression of the adjacent gene, as compared to the wild-type gene. g, Heat map of MarkerFinder cell population-specific genes expression from the Fluidigm scRNA-seq data (n = 191 cells) with enriched pathway associated genes (right) and statistically enriched Gene Ontology biological processes (left). h, i, Heat maps (n = 62 cells) of wild-type-driven genes (same genes as in g) (h) or de novo MarkerFinder genes for cellHarmony classified Gfi1R412X/− cell populations (i) annotated with enriched pathway associated genes (right) and statistically enriched Gene Ontology biological processes (left). j, Representation of the scRNA-seq data from k, displaying the fold change in gene expression in Gfi1R412X/− cells compared to wild-type cells in the indicated clusters, with the number of genes up- or downregulated displayed. Each point represents a single gene. k, Heat map of Fluidigm differentially expressed GFI1-target genes (excluding cell-cycle genes), in which each column represents a single cell and each row represents a single gene, with key genes (right) and enriched biological processes (left) indicated. Cytospin data in a are representative of two biological replicates. Data in a are mean ± s.e.m. ****P < 0.0001, two-tailed t-test in a.
Extended Data Fig. 7 Population analysis of 10x Genomics-based scRNA-seq data and label transfer from CITE-seq transcriptome to scATAC-seq cells.
a, Identification of additional cell populations from wild-type scRNA-seq data (ICGS2 unsupervised analysis, male–female CITE-seq datasets). UMAP projections of scRNA-seq data, in which the cell barcodes within the outlined region (top) were analysed for additional heterogeneity through a second ICGS2 analysis (bottom). b, Heat maps of additional CITE-seq captures with cell assignments from cellHarmony. cellHarmony classifications were derived using the refined cluster annotation assignments from a and Extended Data Fig. 6c. Each panel displays a scRNA-seq heat map of MarkerFinder genes from the cellHarmony reference (top), in which each column represents a single cell and each row represents a single gene. Cell barcodes captured from an independent FACS sort of LY6GhighCD11bhigh GMP-P cells are indicated by a black bar (middle). Relative expression of median-normalized ADTs (right) are shown in the bottom heat maps. c, Assigned cell-population frequencies for Gfi1+/− and Gfi1R412X/− CITE-seq (modified GMP gate) cells data sets from cellHarmony. d, Heat maps of differential ADT expression compared to Gfi1+/− for the indicated markers (right). e, t-distributed stochastic neighbour embedding (t-SNE) plot of CITE-seq transcriptome for the Gfi1+/+ sample. f, t-SNE plot of scATAC-seq for the Gfi1+/+ sample. g, t-SNE plot of CITE-seq transcriptome for the Gfi1R412X/R412X sample. h, t-SNE plot of scATAC-seq for the Gfi1R412X/R412X sample. i, Comparison of Seurat-transferred CITE-seq labels to unsupervised scATAC-seq cell population prediction methods (SNAP-ATAC) for Gfi1+/+. Percentage of overlapping cells for all pairwise comparisons between SNAP-ATAC clusters to CITE-seq clusters derived from Seurat label transfer. j, Comparison of Seurat-transferred CITE-seq labels to unsupervised scATAC-seq cell population prediction methods (SNAP-ATAC) for Gfi1R412X/R412X. Percentage of overlapping cells for all pairwise comparisons between SNAP-ATAC clusters to CITE-seq clusters derived from Seurat label transfer. k, Cell-cluster combined scATAC-seq marker peaks associated with Gfi1+/+ CITE-seq annotated cell populations (Seurat 3 label transfer using cicero gene activity scores). Each row is a cluster (left) and each column is a locus within 50 kb of a CITE-seq marker gene for each cluster (bottom), in which the coloured bars represent the normalized read count coverage. l, Heat maps of transcription factor-motif-enrichment probabilities (−log10-transformed) in accessible regions of Gfi1+/+ (blue) or Gfi1R412X/ R412X (red) cell populations (indicated left). Each row represents a transcription factor motif from the Cisbp2 database (indicated right). Each dot in a and e–h represents a single cell barcode that is pseudo-coloured for its ICGS2 predicted cluster.
Extended Data Fig. 8 In vitro DNA-binding analysis and in vivo ChIP–seq and scATAC-seq analysis of GFI1(R412X).
a–e, EMSA using nuclear extracts of 293T cells transfected with a GFI1, GFI1(R412X), or empty expression vector. Arrows indicate bound (shift) and free probe. All data represent one experiment. a–c, A probe containing a high-affinity GFI1-binding site (R21) was incubated with a titration of nuclear extracts (a) or a titration of cold competitors with (R21) or without (MutR21) a high-affinity GFI1-binding site (b, c).d, e, Nuclear extracts were incubated with probes containing a GFI1-binding site taken from the Mmp8 (d) or Cbx3 (e) locus. These loci contain a GFI1 ChIP–seq peak and the corresponding genes are differentially expressed in Gfi1R412X/− Fluidigm scRNA-seq data. f, Heat map of ChIP–seq read coverage at GFI1-specific peaks, in which each row represents one peak that was called in the wild-type and each column represents a DNA-base position of the peak centred on each GFI1 called peak in the indicated ChIP samples. The cluster colour (left) indicates loci bound by GFI1 alone (white) or by GFI1 and GFI1(R412X) (black). g–l, Schematics of the indicated genomic loci displaying scATAC-seq pseudobulk accessibility in the colour-coded clusters (top) or ChIP–seq reads and called peaks (bottom), with differentially accessible regions bound by GFI1 shaded.
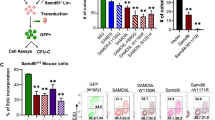
Extended Data Fig. 9 Fluidigm-based transcriptional analysis of Gfi1R412X/− genetic rescues.
a, b, FACS plots (a) and representative cytospins (b) of bone marrow from adult mouse. c, d, Graphical summary of CFU assays performed on KIT+ bone marrow cells. e, FACS plots of mouse bone marrow cells isolated from Irf8-eGFP transgenic mice with the mean percentage of IRF8–GFPhigh cells indicated. f, Three-dimensional FACS plots of bone marrow isolated from adult Irf8-eGFP mice. Events are pseudocoloured for CD11b expression and arrows indicate changes in the incidence of granulocytic populations. g, FACS plots and representative cytospins of adult mouse bone marrow populations sorted for scRNA-seq. h, i, cellHarmony-assigned Gfi1R412X/R412X and Gfi1R412X/−Irf8+/− cells to wild-type reference Fluidigm scRNA-seq populations (h, n = 86 cells, i, n = 88 cells). Each tick mark represents a single-cell library. ICGS clusters are annotated (top). FACS gates are annotated (right). j, Population distribution of Fluidigm scRNA-seq data from h, i and Extended Data Figs. 4c, 6c. j, k, Fluidigm scRNA-seq heat maps (left) of genes that are genetically repaired in each cluster, with key genes indicated. Adjacent plot (right) of enriched biological processes in the indicated clusters. Scale bars, 10 μm. Cytospin data in b are representative of three biological replicates. Data are mean ± s.e.m. (a, c, d) or mean (e). ****P < 0.0001, two-tailed t-test.
Extended Data Fig. 10 G-CSF rescued neutrophils from patients with SCN are morphologically normal but functionally defective.
Representative cytospins of purified human peripheral blood neutrophils. Data are representative of one experiment per donor. Scale bars, 10 μm.
Supplementary information
Supplementary Information
This file contains the Supplementary Methods.
Supplementary Table 1
Deidentified patient information used to identify GFI1 variants of interest and those SCN patients whose neutrophils were assayed.
Supplementary Table 2
Table summarizing the phenotypes of GFI1 variants and the mouse models of SCN.
Supplementary Table 3
Limits of detection for the ProcartaPlex assay used to quantify the listed cytokines in mouse serum.
Supplementary Table 4
Detailed metrics on the cells obtained from each single-cell experiment.
Source data
Rights and permissions
About this article
Cite this article
Muench, D.E., Olsson, A., Ferchen, K. et al. Mouse models of neutropenia reveal progenitor-stage-specific defects. Nature 582, 109–114 (2020). https://doi.org/10.1038/s41586-020-2227-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41586-020-2227-7
This article is cited by
-
Comparative transcriptomics coupled to developmental grading via transgenic zebrafish reporter strains identifies conserved features in neutrophil maturation
Nature Communications (2024)
-
Expansion of interferon inducible gene pool via USP18 inhibition promotes cancer cell pyroptosis
Nature Communications (2023)
-
Asxl1 deletion disrupts MYC and RNA polymerase II function in granulocyte progenitors
Leukemia (2023)
-
ThPOK is a critical multifaceted regulator of myeloid lineage development
Nature Immunology (2023)
-
CD66b−CD64dimCD115− cells in the human bone marrow represent neutrophil-committed progenitors
Nature Immunology (2022)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.