Abstract
The segmental organization of the vertebral column is established early in embryogenesis, when pairs of somites are rhythmically produced by the presomitic mesoderm (PSM). The tempo of somite formation is controlled by a molecular oscillator known as the segmentation clock1,2. Although this oscillator has been well-characterized in model organisms1,2, whether a similar oscillator exists in humans remains unknown. Genetic analyses of patients with severe spine segmentation defects have implicated several human orthologues of cyclic genes that are associated with the mouse segmentation clock, suggesting that this oscillator might be conserved in humans3. Here we show that human PSM cells derived in vitro—as well as those of the mouse4—recapitulate the oscillations of the segmentation clock. Human PSM cells oscillate with a period two times longer than that of mouse cells (5 h versus 2.5 h), but are similarly regulated by FGF, WNT, Notch and YAP signalling5. Single-cell RNA sequencing reveals that mouse and human PSM cells in vitro follow a developmental trajectory similar to that of mouse PSM in vivo. Furthermore, we demonstrate that FGF signalling controls the phase and period of oscillations, expanding the role of this pathway beyond its classical interpretation in ‘clock and wavefront’ models1. Our work identifying the human segmentation clock represents an important milestone in understanding human developmental biology.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
High-throughput sequencing data, raw sequencing data, raw and normalized count data, and single-cell clustering assignments generated in this study have been deposited are available from NCBI Gene Expression Omnibus (GEO) accession number GSE114186, and can be visualized at https://tinyurl.com/DiazPourquie2019. Source Data corresponding to the following figure panels are available with the paper: Fig. 1b–f, 2a–h, 3a–d, f, h, 4b–i and Extended Data Figs. 1 c, d, f–k, m–s, 2a–c, 3a, b, d–g, i–l, n, o, 4a–d, 5b, c, g–w, 6b, c, f–q, s–u, 7b–d. Online interactive versions and downloadable versions of the analysed scRNA-seq datasets, as well as scRNA-seq transcript × cell count tables can be accessed at https://tinyurl.com/DiazPourquie2019, as follows. Mouse E9.5 t-distributed stochastic neighbour-embedding (t-SNE) clustering analysis (Extended Data Fig. 2c) data are available from https://tinyurl.com/DiazPourquie2019-mE95. The mouse E9.5 k-NN graph of paraxial mesoderm and neural clusters (Fig. 2a, b, Extended Data Fig. 3a–e) is available from: https://tinyurl.com/DiazPourquie2019-mE95-PSM. Data related to mouse ES cell cultures from day 0 to day 5 (Fig. 2c, d, Extended Data Figs. 3f–j) are available from: https://tinyurl.com/DiazPourquie2019-mESC. Data related to human iPS cell cultures from day 0 to day 4 (Fig. 2e, f, Extended Data Figs. 3k–o) are available from: https://tinyurl.com/DiazPourquie2019-hIPSC. Additional data, such a raw image files, are available from the corresponding author upon request; all materials used in this study—including stem cell lines carrying knock-in reporters—are available by request from the corresponding author.
Code availability
Single-cell sequencing data were processed and analysed using publicly available software packages: https://github.com/indrops/indrops and https://github.com/AllonKleinLab/SPRING. Downstream analysis was performed in ScanPy38 (1.4.3), using Python 3.6.8. Python code and Jupyter notebooks for reproducing single-cell analyses appearing in Fig. 2 and Extended Data Figs 2–4 are available at https://github.com/wagnerde/Diaz2019. This Github link also includes detailed instructions for installing the necessary Python software environment, including the following packages and their dependencies: anndata(0.6.22.post1), bbknn(1.3.6), fa2(0.3.5), ipython(7.8.0), jupyterlab(1.1.4), leidenalg(0.7.0), louvain(0.6.1), matplotlib(3.0.3), multicoretsne(0.1), numba(0.45.1), numpy(1.17.2), pandas(0.25.1), pytables(3.5.2), python(3.6.7), python-igraph(0.7.1.post7), scanpy(1.4.4.post1), scikit-learn(0.21.3), scipy(1.3.1), scrublet(0.2.1), seaborn(0.9.0), statsmodels(0.10.1) and umap-learn(0.3.10). Force-directed layouts of single-cell graphs were generated using the ForceAtlas2 algorithm in Gephi (0.9.1). MATLAB code used for single-cell tracking and synchronization analysis is available at: https://github.com/jonathan-touboul-brandeis/HumanSegmentationClock.
Change history
12 February 2024
Editor's Note: Readers are alerted that the reliability of data presented in this article is currently in question due to errors in the cell tracking analysis code. The data is being re-analyzed, and appropriate editorial action will be taken once the full impact of this error is determined.
References
Hubaud, A. & Pourquié, O. Signalling dynamics in vertebrate segmentation. Nat. Rev. Mol. Cell Biol. 15, 709–721 (2014).
Oates, A. C., Morelli, L. G. & Ares, S. Patterning embryos with oscillations: structure, function and dynamics of the vertebrate segmentation clock. Development 139, 625–639 (2012).
Sparrow, D. B. et al. Mutation of the LUNATIC FRINGE gene in humans causes spondylocostal dysostosis with a severe vertebral phenotype. Am. J. Hum. Genet. 78, 28–37 (2006).
Chal, J. et al. Differentiation of pluripotent stem cells to muscle fiber to model Duchenne muscular dystrophy. Nat. Biotechnol. 33, 962–969 (2015).
Hubaud, A., Regev, I., Mahadevan, L. & Pourquie, O. Excitable dynamics and Yap-dependent mechanical cues drive the segmentation clock. Cell 171, 668–682 (2017).
Chal, J. et al. Generation of human muscle fibers and satellite-like cells from human pluripotent stem cells in vitro. Nat. Protocols 11, 1833–1850 (2016).
Henrique, D., Abranches, E., Verrier, L. & Storey, K. G. Neuromesodermal progenitors and the making of the spinal cord. Development 142, 2864–2875 (2015).
Gouti, M. et al. A gene regulatory network balances neural and mesoderm specification during vertebrate trunk development. Dev. Cell 41, 243–261 (2017).
Klein, A. M. et al. Droplet barcoding for single-cell transcriptomics applied to embryonic stem cells. Cell 161, 1187–1201 (2015).
Tzouanacou, E., Wegener, A., Wymeersch, F. J., Wilson, V. & Nicolas, J. F. Redefining the progression of lineage segregations during mammalian embryogenesis by clonal analysis. Dev. Cell 17, 365–376 (2009).
Chalamalasetty, R. B. et al. Mesogenin 1 is a master regulator of paraxial presomitic mesoderm differentiation. Development 141, 4285–4297 (2014).
Matsumiya, M., Tomita, T., Yoshioka-Kobayashi, K., Isomura, A. & Kageyama, R. ES cell-derived presomitic mesoderm-like tissues for analysis of synchronized oscillations in the segmentation clock. Development 145, dev156836 (2018).
Yoshioka-Kobayashi, K. et al. Coupling delay controls synchronized oscillations in the segmentation clock. Nature https://doi.org/10.1038/s41586-019-1882-z (2019).
Aulehla, A. et al. A β-catenin gradient links the clock and wavefront systems in mouse embryo segmentation. Nat. Cell Biol. 10, 186–193 (2008).
Masamizu, Y. et al. Real-time imaging of the somite segmentation clock: revelation of unstable oscillators in the individual presomitic mesoderm cells. Proc. Natl Acad. Sci. USA 103, 1313–1318 (2006).
Chal, J. et al. Recapitulating early development of mouse musculoskeletal precursors of the paraxial mesoderm in vitro. Development 145, dev157339 (2018).
Bénazéraf, B. et al. A random cell motility gradient downstream of FGF controls elongation of an amniote embryo. Nature 466, 248–252 (2010).
Acebrón, J. A., Bonilla, L. L., Perez Vicente, C. J., Ritort, F. & Spigler, R. The Kuramoto model: a simple paradigm for synchronization phenomena. Rev. Mod. Phys. 77, 137–185 (2005).
Palmeirim, I., Henrique, D., Ish-Horowicz, D. & Pourquié, O. Avian hairy gene expression identifies a molecular clock linked to vertebrate segmentation and somitogenesis. Cell 91, 639–648 (1997).
Primmett, D. R., Norris, W. E., Carlson, G. J., Keynes, R. J. & Stern, C. D. Periodic segmental anomalies induced by heat shock in the chick embryo are associated with the cell cycle. Development 105, 119–130 (1989).
Tsiairis, C. D. & Aulehla, A. Self-organization of embryonic genetic oscillators into spatiotemporal wave patterns. Cell 164, 656–667 (2016).
Jiang, Y. J. et al. Notch signalling and the synchronization of the somite segmentation clock. Nature 408, 475–479 (2000).
Riedel-Kruse, I. H., Muller, C. & Oates, A. C. Synchrony dynamics during initiation, failure, and rescue of the segmentation clock. Science 317, 1911–1915 (2007).
Dupont, S. et al. Role of YAP/TAZ in mechanotransduction. Nature 474, 179–183 (2011).
Cooke, J. & Zeeman, E. C. A clock and wavefront model for control of the number of repeated structures during animal morphogenesis. J. Theor. Biol. 58, 455–476 (1976).
Morimoto, M., Takahashi, Y., Endo, M. & Saga, Y. The Mesp2 transcription factor establishes segmental borders by suppressing Notch activity. Nature 435, 354–359 (2005).
Hauschka, S. D. Clonal analysis of vertebrate myogenesis. II. Environmental influences upon human muscle differentiation. Dev. Biol. 37, 329–344 (1974).
Ran, F. A. et al. Genome engineering using the CRISPR–Cas9 system. Nat. Protocols 8, 2281–2308 (2013).
Oceguera-Yanez, F. et al. Engineering the AAVS1 locus for consistent and scalable transgene expression in human iPSCs and their differentiated derivatives. Methods 101, 43–55 (2016).
Wang, H. et al. NOTCH1-RBPJ complexes drive target gene expression through dynamic interactions with superenhancers. Proc. Natl Acad. Sci. USA 111, 705–710 (2014).
Morales, A. V., Yasuda, Y. & Ish-Horowicz, D. Periodic Lunatic fringe expression is controlled during segmentation by a cyclic transcriptional enhancer responsive to notch signaling. Dev. Cell 3, 63–74 (2002).
Hayashi, S., Nakahata, Y., Kohno, K., Matsui, T. & Bessho, Y. Presomitic mesoderm-specific expression of the transcriptional repressor Hes7 is controlled by E-box, T-box, and Notch signaling pathways. J. Biol. Chem. 293, 12167–12176 (2018).
Schindelin, J. et al. Fiji: an open-source platform for biological-image analysis. Nat. Methods 9, 676–682 (2012).
Delaune, E. A., François, P., Shih, N. P. & Amacher, S. L. Single-cell-resolution imaging of the impact of Notch signaling and mitosis on segmentation clock dynamics. Dev. Cell 23, 995–1005 (2012).
Goldberg, J. M. & Brown, P. B. Response of binaural neurons of dog superior olivary complex to dichotic tonal stimuli: some physiological mechanisms of sound localization. J. Neurophysiol. 32, 613–636 (1969).
Gordon, A. Y. & Lev B. K. in Nonparametrics and Robustness in Modern Statistical Inference and Time Series Analysis: A Festschrift in Honor of Professor Jana Jurečková (eds Antoch, J. et al.) 70–74 (Institute of Mathematical Statistics, 2010).
Zilionis, R. et al. Single-cell barcoding and sequencing using droplet microfluidics. Nat. Protocols 12, 44–73 (2017).
Wolf, F. A., Angerer, P. & Theis, F. J. SCANPY: large-scale single-cell gene expression data analysis. Genome Biol. 19, 15 (2018).
Wolock, S. L., Lopez, R. & Klein, A. M. Scrublet: computational identification of cell doublets in single-cell transcriptomic data. Cell Syst. 8, 281–291.e9 (2019).
Polański, K. et al. BBKNN: fast batch alignment of single cell transcriptomes. Bioinformatics https://doi.org/10.1093/bioinformatics/btz625 (2019).
Blondel, V. D. et al. Fast unfolding of communities in large networks. J. Stat. Mech. 2008, P10008 (2008).
Traag, V. A., Waltman, L. & van Eck, N. J. From Louvain to Leiden: guaranteeing well-connected communities. Sci. Rep. 9, 5233 (2019).
Macosko, E. Z. et al. Highly parallel genome-wide expression profiling of individual cells using nanoliter droplets. Cell 161, 1202–1214 (2015).
Wolf, F. A. et al. PAGA: graph abstraction reconciles clustering with trajectory inference through a topology preserving map of single cells. Genome Biol. 20, 59 (2019).
Loh, K. M. et al. Mapping the pairwise choices leading from pluripotency to human bone, heart, and other mesoderm cell types. Cell166, 451–467 (2006).
Zhou, H. et al. Conversion of mouse epiblast stem cells to an earlier pluripotency state by small molecules. J. Biol. Chem. 285, 29676–29680 (2010).
Du, W., Prochazka, J., Prochazkova, M. & Klein, O. D. Expression of FGFs during early mouse tongue development. Gene Exp. Patterns 20, 81–87 (2016).
Acknowledgements
We thank members of the Pourquié laboratory, D. Ish-Horowicz, A. Klein and M. Heiman for critical reading of the manuscript and discussions. Research in the Pourquié laboratory was funded by a grant from the National Institute of Health (5R01HD085121). D.E.W. is supported by 1K99GM121852. M.D.-C. is supported by the NSF GRFP.
Author information
Authors and Affiliations
Contributions
M.D.-C. designed, performed and analysed biological experiments with O.P.; D.E.W. analysed scRNA-seq data. C.B. optimized the dissociation protocol for scRNA-seq and contributed to experiments with M.D.-C. A.H. performed mouse explant experiments. O.A.T. performed ChIP–qPCR experiments and helped M.D.-C. and S.D. to generate the mouse Hes7-Achilles line. S.D. helped M.D.-C. to generate the mouse Hes7-Achilles line and the human HES7-Achilles;pCAG-H2B-mCherry and HES7-Achilles;MESP2-mCherry lines. A. Michaut. helped with the quantifications. Z.A.T. generated the MSGN1-YFP line and helped M.D.-C. with generation of the HES7-Achilles line. K.Y.-K. and R.K. generated the destabilized Achilles construct. Y.N. and A. Miyawaki. generated the Achilles protein. J.T. performed the automated cell tracking and mathematical analysis of synchronization. M.D.-C., D.E.W., A.H., C.B., J.T. and O.P. wrote the manuscript; and O.P. supervised the project. All authors discussed and agreed on the results and commented on the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare the following competing interests: O.P. is scientific founder of Anagenesis Biotechnologies.
Additional information
Peer review information Nature thanks Helen M. Blau and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
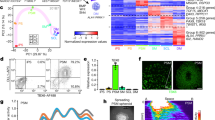
Extended Data Fig. 1 Differentiation of mouse and human pluripotent stem cells towards PSM fate for the characterization of the segmentation clock in vitro.
a, Scheme illustrating the maturation stages of paraxial mesoderm. DF, determination front; pPSM. b, Top, immunofluorescence staining for the cadherins CDH1 and CDH2 (top), and the pluripotency factor POU5F1 (bottom) in differentiating mouse ES cells (ESCs) (left) and human iPS cells (iPSCs) (right). n = 4 independent experiments. Scale bar, 100 μm. c, qRT–PCR for the epiblast marker Fgf5, the neuromesodermal progenitor or mesodermal marker T, and the mesodermal precursor cell and PSM markers Tbx6, Msgn1 and Rspo3 on days 2–6 of mouse ES cell differentiation. Relative expression is shown as the fold change relative to ES cells at day 0. Mean ± s.d. n = 3 biological replicates. d, Percentage induction of the mouse (m)ES cell pMsgn1-Venus reporter and the human (h)iPS cell MSGN1-Venus reporter, as determined by fluorescence-activated cell sorting (FACS). Mean ± s.d. n = 12 independent experiments (mouse ES cell), n = 8 independent experiments (human iPS cell). e, Gating strategy and representative FACS plots for quantification of pMsgn1-Venus or MSGN1-Venus induction. f, qRT–PCR for cyclic genes (HES7 and LFNG), posterior-PSM markers (MSGN1, TBX6 and RSPO3), determination-front markers (MESP2 and RIPPLY2) and anterior-PSM markers (MEST and FOXC2) on days 1–4 of human iPS cell (iPSC) differentiation. Relative expression is shown as the fold change relative to iPS cells at day 0. Mean ± s.d. n = 3 biological replicates. g, Diagram outlining the targeting strategy used to generate Hes7-Achilles and HES7-Achilles knock-in reporter lines in mouse ES cells and human iPS cells, respectively. h, Normalized HES7–Achilles fluorescence intensity for three PSM cells derived from mouse ES cells, imaged in CL medium on day 4 of differentiation. n = 4 independent experiments. i, Representative Fourier transform of HES7–Achilles oscillations in PSM cells derived from mouse ES cells, indicating the predominant period. n = 19 cells. j, Total time spent in the oscillatory state for Hes7-Achilles PSM cells derived from mouse ES cells, cultured in CL or CLFBR medium from day 4 onwards. The middle hinge corresponds to median, the lower and upper hinges correspond to the first and third quartiles, respectively, and the lower and upper whiskers correspond to the minimum and maximum, respectively. n = 8 (CL), n = 12 (CLFBR) independent experiments. k, qRT–PCR comparing relative expression levels of Msgn1, Lfng, T and Tbx6 in PSM cells derived from mouse ES cells, cultured in CL or CLFBR medium from day 4 onwards. Relative expression is shown as the fold change relative to ES cells at day 0. Mean ± s.d. n = 3 biological replicates. l, Snapshots of HES7–Achilles fluorescence in PSM cells derived from human iPS cells, showing peaks and troughs over the course of 13.5 h in CL medium on day 2 of differentiation. n = 25 independent experiments. Scale bar, 100 μm. m, Representative quantification of HES7–Achilles fluorescence intensity in a small ROI from day 2 to day 3 of human iPS cell differentiation. n = 25 independent experiments. n, Representative Fourier transform of HES7-Achilles oscillations, indicating the predominant period in PSM cells derived from human iPS cells, in CL medium on day 2. n = 25 independent experiments. o, Representative instantaneous frequency in Hertz (calculated by Hilbert transformation) of HES7-Achilles oscillations in PSM cells derived from human iPS cells, from day 2 to day 3 of differentiation in CL medium. n = 25 independent experiments. p, Representative instantaneous frequency in Hertz (calculated by Hilbert transformation) of HES7-Achilles oscillations in PSM cells derived from human iPS cells, from day 2 to day 3 of differentiation in CLFBR medium. n = 33 independent experiments. q, Quantification of HES7–Achilles fluorescence in human iPS cells differentiated for 48 h without the BMP inhibitor LDN93189 (CHIR99021-only medium). n = 3 independent experiments. r, Total number of HES7-Achilles oscillations for PSM cells derived from human iPS cells, cultured in CL or CLFBR medium from day 2 onwards. Mean ± s.d. n = 15 independent experiments. s, qRT–PCR comparing relative expression levels of HES7, LFNG, TBX6 and MSGN1 in PSM cells derived from human iPS cells, cultured in CL or CLFBR medium from day 2 onwards. Relative expression is shown as the fold change relative to iPS cells on day 0. Mean ± s.d. n = 3 biological replicates.
Extended Data Fig. 2 scRNA-seq analysis of the mouse E9.5 embryonic tail bud.
a, Prefiltering of doublet-like cells. UMAP embedding shows all original E9.5 cells (n = 5,646), coloured by doublet score. Doublet scores indicate the extent to which a given single-cell transcriptome resembles a linear combination of two randomly selected cells (Methods and ref. 39). b, Histogram of doublet scores. Scores >0.24 were filtered from subsequent analyses. c, t-SNE embedding of E9.5 cells (n = 4,367) post-doublet filtering. Individual cells are coloured according to annotated Louvain cluster identities. d, Top 20 positively enriched transcripts for each Louvain cluster relative to all other clusters, as detected by a two-sided Wilcoxon rank-sum test. Reported transcripts are ranked by FDR-corrected P values (Benjamini–Hochberg). Exact sample sizes are given in Supplementary Table 1.
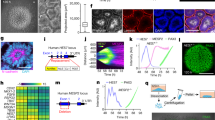
Extended Data Fig. 3 Comparative analysis of PSM differentiation trajectories in vitro and in vivo.
a, f, k, ForceAtlas2 layouts of mouse E9.5 embryos, mouse ES cell and human iPS cell single-cell k-NN graphs, coloured by cluster identity and collection time points, as indicated. b, g, l, Confusion matrices plot the overlap of cluster and time-point assignments, row-normalized. c, h, m, Top 20 positively enriched transcripts for Louvain clusters relative to all other clusters in each dataset, as detected by a two-sided Wilcoxon rank-sum test. Reported transcripts are ranked by FDR-corrected P values (Benjamini–Hochberg). Exact sample sizes are given in Supplementary Table 1. d, i, n, ForceAtlas2 layouts of single-cell k-NN graphs, overlaid with log-normalized transcript counts for indicated genes. e, j, o, Top, colours indicate pseudotemporal orderings. Bottom, heat map of selected markers of paraxial mesoderm differentiation. Approximate locations of cluster centres are indicated.
Extended Data Fig. 4 Fate outcomes of PSM-directed differentiation in human and mouse-derived cultures.
a, c, ForceAtlas2 layouts of indicated single-cell k-NN graphs, overlaid with classifier prediction scores. b, d, Heat map of single-cell HOX-gene expression levels for mouse ES cell and human iPS cell datasets. Columns (individual cells) are grouped by collection time point. Rows are individual HOX genes ordered by position. Approximate anatomical positions of HOX paralogues are indicated on the right.
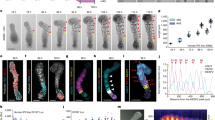
Extended Data Fig. 5 Analysis of the human segmentation clock at the single-cell level.
a, Scheme showing the insertion of a constitutively expressed pCAG-H2B-mCherry nuclear label in the safe harbour AAVS1 locus in a HES7-Achilles human-iPS-cell background. b, Diffusion (square micrometres per minute) for individual human HES7-Achilles cells automatically tracked over a period of 24 h. The middle hinge corresponds to median, lower and upper hinges correspond to first and third quartiles, respectively, and the lower and upper whiskers correspond to the minimum and maximum, respectively. n = 76 cells. c, Distribution of pairwise instantaneous phase shifts between individual oscillating human HES7-Achilles cells, binned by instantaneous distance between pairs of cells. P values for the pairwise Kolmogorov–Smirnov test are as follows: <160 μm versus 160–265 μm: 0.6407, <160 μm versus 265–530 μm: 0.1811, <160 μm versus >530 μm: 0.1340, 160–265 μm versus 265–530 μm: 0.1428, 160–265 μm versus >530 μm: 0.6784, and 265–530 μm versus >530 μm: 0.8171. n = 1,000 observations. d, Distribution of phases along the unit circle at early, middle and late time points. Each dot represents one cell. n = 144 cells. e, Illustration of phase determination. Representative raw HES7–Achilles fluorescence profile for an automatically tracked cell (left) and corresponding processed signal along with the inferred phase from Hilbert transform (right). f, Heat map of HES7–Achilles fluorescence intensity over time in automatically tracked cells. Each line represents one cell. n = 144 cells. g, Histogram of the time (hours since onset of imaging) at cell division for manually tracked human HES7-Achilles cells. n = 67 cells. h, Left, immunofluorescence staining for histone H3 phosphorylated at Ser10, in PSM cells derived from human iPS cells, treated with vehicle control (DMSO) or 5 μM aphidicolin for 24 or 48 h, starting on day 2 of differentiation. n = 5 independent experiments. Scale bar, 100 μm. Right, quantification of phosphorylated histone H3 (at Ser10) nuclei as a percentage of total nuclei. The middle hinge corresponds to median, the lower and upper hinges correspond to the first and third quartiles, respectively, and the lower and upper whiskers correspond to the minimum and maximum, respectively. i, Scatter plot showing the cell-cycle time in PSM cells derived from human iPS cells, cultured in CLFBR medium. Mean ± s.d. n = 26 cells. j, Histogram of the HES7–Achilles oscillatory phase at the time of cell division in human iPSC-derived PSM cells cultures in CLFBR medium. Distribution is not significantly different from the uniform distribution: Kolmogorov-Smirnov test p=0.225. n=55 cell divisions. k, Normalized HES7–Achilles fluorescence intensity profiles for 3 individual PSM cells derived from human iPS cells, pre-treated with 5 μM Aahidicolin for 24 h. n = 6 independent experiments. l, Kuramoto order parameter over 20 h on day 3 of differentiation for human HES7-Achilles cells treated with vehicle control (DMSO) or 5 μM aphidicolin for 24 h. The synchronization threshold is shown as the mean ± s.d. of the Kuramoto order parameter for same dataset, but with randomized phases. n = 45 cells (control) or 48 cells (aphidicolin). m, Comparison of the Kuramoto order parameter for oscillating HES7-Achilles cells treated with vehicle control (DMSO) or 5 μM aphidicolin. The middle hinge corresponds to the median, the lower and upper hinges correspond to the first and third quartiles, respectively, and the lower and upper whiskers correspond to the minimum and maximum, respectively. Paired two-sided t-test, P = 0.348. n = 45 cells (control) or 48 cells (aphidicolin). n, qRT–PCR for Notch target genes HES7, NRARP and LFNG in PSM cells derived from human iPS cells, treated with vehicle control (DMSO) or 25 μM DAPT on day 2 of differentiation. Mean ± s.d. n = 3 biological replicates. o, Example of HES7–Achilles fluorescence intensity in a small ROI over a period of 45 h in cells treated with DMSO (vehicle control) or the γ-secretase inhibitor DAPT (25 μM) in CLFBR medium. n = 16 independent experiments. p, Representative example of HES7–Achilles fluorescence intensity profiles for PSM cells derived from mouse ES cells, treated with vehicle control (DMSO) or 25 μM DAPT. n = 13 independent experiments. q, Kuramoto order parameter over 20 h on day 2 of differentiation for human HES7-Achilles cells treated with vehicle control (DMSO) or 25 μM DAPT. The synchronization threshold is shown as the mean ± s.d. of the Kuramoto order parameter for same dataset, but with randomized phases. n = 131 cells (control) or 110 cells (DAPT). r, Representative immunofluorescence staining for YAP, F-actin (phalloidin) and DAPI nuclear stain in isolated human PSM-like cells treated with DMSO or latrunculin A (350 nM). Scale bar, 50 μm. n = 4 independent experiments. s, ChIP–qPCR fold enrichment of the LFNG and HES7 promoters in chromatin pulled down with an antibody against NOTCH1, relative to isotype IgG controls. Mean ± s.d. iPS-cell control, n = 4; all other conditions, n = 3 biological replicates. t, Mean HES7–Achilles fluorescence intensity for isolated human cells cultured with 350 nM latrunculin A alone, or in combination with 25 μM DAPT. The middle hinge corresponds to the median, the lower and upper hinges correspond to the first and third quartiles, respectively, and the lower and upper whiskers correspond to the minimum and maximum, respectively. n = 18 cells. u, Scatter plot showing the HES7–Achilles oscillatory period for isolated human cells cultured with 350 nM latrunculin A alone, or in combination with 25 μM DAPT. Mean ± s.d. n = 47 (latrunculin A) or 22 (latrunculin A + DAPT) cells. v, Kuramoto order parameter over 18 h on day 2 of differentiation for human HES7-Achilles cells treated with DMSO, latrunculin A alone or latrunculin A in combination with DAPT. The synchronization threshold is shown as the mean ± s.d. of the Kuramoto order parameter for the same dataset, but with randomized phases. n = 53 cells (control), 18 cells (latrunculin A) or 18 cells (latrunculin A + DAPT). w, Comparison of the Kuramoto order parameter in confluent HES7-Achilles cells versus isolated cells treated with 350 nM latrunculin A alone, or in combination with 25 μM DAPT. The middle hinge corresponds to the median, the lower and upper hinges correspond to the first and third quartiles, respectively, and the lower and upper whiskers correspond to the minimum and maximum, respectively. Paired one-way ANOVA with Bonferroni correction: confluent control versus LatA, P = 1.16 × 10−6; confluent control versus latrunculin A + DAPT, P = 6.8 × 10−13; latrunculin A versus latrunculin A + DAPT, P = 0.304. n = 53 cells (control), 18 cells (latrunculin A) or 18 cells (latrunculin A + DAPT).
Extended Data Fig. 6 The role of FGF and WNT signaling in the regulation of segmentation clock properties.
a, Immunofluorescence staining for dpERK on day 2 of differentiation, following 3 h of treatment with DMSO (vehicle control) or the FGFR inhibitor PD173074 (250 nM) in CL medium. n = 4 independent experiments. b, Left, qRT–PCR for the FGF ligands FGF17 and FGF8 on days 1–4 of human iPS cell differentiation. Relative expression is shown as the fold change relative to iPS cells at day 0. Mean ± s.d. n = 3 biological replicates. Right, qRT–PCR for the FGF ligand Fgf8 on days 2–6 of mouse ES cell differentiation. Relative expression is shown as the fold change relative to ES cells at day 0. Mean ± s.d. n = 3 biological replicates. c, Time-course qRT–PCR for the FGF target gene SPRY4 in PSM cells derived from human iPS cells, during the 10 h immediately after treatment with vehicle control (DMSO) or 250 nM PD03. Relative expression is shown as the fold change relative to ES cells at day 0. Mean ± s.d. n = 3 biological replicates. d, Immunofluorescence staining for dpERK in PSM cells derived from human iPS cells (top) or from mouse ES cells (bottom), treated with DMSO or PD03 (250 nM). n = 4 independent experiments. e, Immunofluorescence staining for dpERK (left), β-catenin and nuclear stain (right) in PSM cells derived from human iPS cells, treated with vehicle control (DMSO) or 2 μM XAV. n = 4 independent experiments. Scale bar, 100 μm. f, Representative examples of HES7–Achilles fluorescence intensity over the course of 45 h in a small area of interest within human cultures treated with DMSO (vehicle control), the MAPK inhibitor PD0325901 (250 nM), or the FGFR inhibitor PD173074 (250 nM) in CLFBR medium. n = 16 independent experiments. g, HES7–Achilles fluorescence intensity over the course of 45 h in a small ROI within human cultures treated with DMSO (vehicle control), 2 μM XAV or 12 μM IWR-1 in CLFBR medium. n = 3 independent experiments. h, HES7-Achilles oscillatory period of individual cells treated with vehicle control (DMSO), 2 μM XAV, 250 nM PD17, or 100 nM, 250 nM or 500 nM PD03 on day 2 of differentiation. Mean ± s.d. One-way ANOVA P values (NS, not significant): 0.9929, 0.4097, 0.9998, 0.9845 and 0.7425, from left to right on the graph. n = 27 (XAV), n = 48 (100 nM PD03), n = 57 (all others) cells. i, Average fluorescence intensity profiles for automatically tracked individual HES7-Achilles human cells treated with vehicle control (DMSO) or increasing doses of PD03 (100 nM, 250 nM and 500 nM) on day 2 of differentiation. Mean ± 95% confidence interval. n = 68 cells (control), 45 cells (100 nM), 35 cells (250 nM) or 36 cells (500 nM). j, Representative examples of HES7–Achilles fluorescence intensity profiles in a small ROI within human cultures treated with increasing doses of PD03 (100 nM, 250 nM and 500 nM) or vehicle control (DMSO). n = 8 independent experiments. k, Number of HES7–Achilles oscillations before arrest in small ROIs within cultures treated with increasing doses of PD03 (100 nM, 250 nM and 500 nM). One-way ANOVA: 100 nM versus 250 nM, P = 0.0042; 100 nM versus 500 nM, P = 2.0 × 10−5. n = 6 independent experiments. l, Average HES7–Achilles fluorescence intensity in small ROIs over the course of the oscillatory regime (before the arrest of oscillations) in cells treated with vehicle control (DMSO) or increasing doses of PD03 (100 nM, 250 nM and 500 nM). The middle hinge corresponds to the median, the lower and upper hinges correspond to the first and third quartiles, and the lower and upper whiskers correspond to the minimum and maximum. One-way ANOVA: control versus 100 nM, P = 6.7 × 10−17; control versus 250 nM, P = 6.5 × 10−21; control versus 500 nM, P = 1.9 × 10−22; 100 nM versus 250 nM, P = 1.1 × 10−17; 100 nM versus 500 nM, P = 2.5 × 10−18. n = 6 independent experiments. m, Representative HES7–Achilles fluorescence intensity profiles for PSM cells derived from mouse ES cells, treated with vehicle control (DMSO), 2 μM XAV, or 100 nM, 250 nM or 500 nM PD03. n = 12 (control, XAV and 100 nM PD03) or n = 10 (250 nM and 500 nM PD03) independent experiments. n, Histograms showing the instantaneous phase difference relative to control for individual cells treated with vehicle control (DMSO), 2 μM XAV, 250 nM PD17, or 100 nM, 250 nM or 500 nM PD03. Details are given in ‘Phase shifts’ in Methods. n was fixed at 11,000 observations. o, Quantification of the average phase difference (in degrees) for HES7–Achilles oscillations in small ROIs in cells treated with 250 nM PD17, or 100 nM, 250 nM or 500 nM PD03 relative to control (DMSO) cells. The middle hinge corresponds to the median, the lower and upper hinges correspond to the first and third quartiles, respectively, and the lower and upper whiskers correspond to the minimum and maximum, respectively. n = 13 (PD17), n = 17 (100 nM), n = 7 (250 nM) or n = 11 (500 nM) independent experiments. p, q, Time-lapse qRT–PCR for the cyclic genes HES7 (p) and LFNG (q) in PSM cells derived from human iPS cells, under control (DMSO) and 250-nM PD03 conditions. Samples were taken every 30 min immediately after treatment. Relative expression is shown as the fold change relative to ES cells at day 0. Mean ± s.d. n = 3 technical replicates. r, Outline of the experimental strategy used to assess the effect of FGF inhibition in primary mouse PSM cells carrying the LuVeLu reporter. The tail bud is dissected from E9.5 transgenic embryos, and cells are dissociated for seeding on fibronectin micropatterns. Oscillations of the LuVeLu reporter are examined in each micropattern. s, LuVeLu fluorescence intensity profiles in mouse tail-bud explant cells cultured on CYTOO micropatterns in CLFBR medium containing DMSO (vehicle control) or increasing doses of PD03 (0.4 μM, 0.65 μM and 10 μM). n = 2 independent experiments. t, Number of LuVeLu oscillations before arrest in mouse tail-bud explant cells cultured on CYTOO micropatterns treated with DMSO (vehicle control) or increasing doses of PD03 (0.4 μM, 0.65 μM and 10 μM). Mean ± s.d. One way ANOVA: 0.4 μM versus 0.65 μM, P = 0.0642; 0.4 μM versus 10 μM, P = 8.4 × 10−9; 0.65 μM versus 10 μM, P = 2.9 × 10−6. n = 10 micropatterns (0.4 μM), n = 7 micropatterns (0.65 μM and 10 μM) u, Average period of LuVeLu oscillations in mouse tail-bud explant cells cultured on CYTOO micropatterns treated with DMSO (vehicle control) or increasing doses of PD03 (0.4 μM, 0.65 μM and 10 μM). Mean ± s.d. One way ANOVA: control versus 0.4 μM, P = 0.2785; control versus 0.65 μM, P = 0.0658; control versus 10 μM, P = 2.7 × 10−6; 0.4 μM versus 0.65 μM, P = 0.831; 0.4 μM versus 10 μM, P = 3.05 × 10−4; 0.65 μM versus 10 μM, P = 4 × 10−3. n = 18 micropatterns (DMSO), n = 16 micropatterns (0.4 μM), n = 12 micropatterns (0.65 μM) and n = 6 micropatterns (10 μM).
Extended Data Fig. 7 Control of PSM maturation by FGF and WNT signalling in vitro.
a, Snapshots of HES7-Achilles;MESP2-mCherry double-reporter cells on days 2–5 of differentiation in CLFBR medium at 0, 20, 45 and 60 h. Cultures treated with DMSO (control), 25 μM DAPT, 2 μM XAV and 250 nM PD03 are shown. n = 10 independent experiments. Scale bar, 100 μm. b, Time of onset of MESP2-mCherry expression in PSM cells derived from human iPS cells treated with vehicle control (DMSO), 2 μM XAV, 250 nM PD17 or 100 nM, 250 nM or 500 nM PD03. Onset of expression is defined by a threshold of 25 AU. Mean ± s.d. One-way ANOVA: control versus XAV, P = 4.6 × 10−15; control versus 100 nM PD03, P = 5.1−17; control versus 250 nM PD03, P = 1.3 × 10−17; control versus 500 nM PD03, P = 1.4 × 10−18; 100 nM versus 250 nM PD03, P = 2.6 × 10−15; 100 nM versus 500 nM PD03, P = 7.7 × 10−16; 250 nM versus 500 nM PD03, P = 6.9 × 10−5. n = 10 independent experiments. c, HES7–Achilles and MESP2–mCherry fluorescence intensity profiles in small ROIs within PSM cultures derived from human iPS cells, treated with 25 μM DAPT on days 2–5 of differentiation in CLFBR medium. Mean ± s.d. Dotted line denotes the threshold for MESP2 activation (25 AU). n = 15 independent experiments. d, qRT–PCR for the genes HES7, LFNG, MSGN1, TBX6, DUSP6, FOXC2, MESP2 and RIPPLY2 in PSM cultures derived from human iPS cells, treated for 24 h with vehicle control (DMSO) or 250 nM PD03 in CLFBR medium. Relative expression is shown as the fold change relative to iPS cells at day 0. Mean ± s.d. n = 3 biological replicates. e, Immunofluorescence staining for TBX6 on day 3 of differentiation (CLFBR medium) in cells treated with DMSO or PD03 (250 nM). n = 4 independent experiments. Scale bar, 100 μm.
Supplementary information
Supplementary Table
Supplementary Table 1: Differentially expressed genes for cell groups defined by Louvain clustering, determined by a two-sided Wilcoxon Rank-Sum test relative to cells of all other clusters in a given dataset. Up to 100 enriched gene names and associated fold changes are listed for each cluster, ranked by Benjamini-Hochberg-corrected p-value (adj. p < 0.05). Results are reported for cell clusters appearing in Extended Data Fig. 2a (Mouse E9.5), Fig. 2a (Mouse E9.5 PSM), Fig. 2e (Human iPSC), and Fig. 2c (Mouse ESC). Sample sizes (i.e. number of cells) for each cluster are also reported, and range from n=24 to n=7347.
Supplementary Table
Supplementary Table 2: Table of significant dynamically expressed genes along the paraxial mesoderm cell state trajectory of the mouse E9.5 embryo (see Fig. 2b and Methods). Listed for each gene are p-value (FDR-corrected), the trajectory-ordered cell (n=1410) with peak expression, and gene order.
41586_2019_1885_MOESM4_ESM.mp4
Video 1: mHes7-Achilles oscillations, related to Fig. 1. Timelapse imaging of mouse ESC-derived Hes7-Achilles PSM cells on days 4-5 of differentiation in CLFBR medium. Saturated pixels corresponding to autofluorescent debris have been computationally removed for clarity (see methods). n=17 independent experiments. Scale bar = 100µm.
Video 2:
Induction of hMSGN1-Venus and EMT, related to Fig. 1. Timelapse imaging of cells carrying the MSGN1-Venus reporter on days 1-2 of differentiation. n=7 independent experiments. Scale bar = 100µm.
Video 3:
hHES7-Achilles oscillations, related to Fig. 1. Timelapse imaging of the human iPSC HES7-Achilles line on days 2-3 of differentiation in CL medium. n=25 independent experiments. Scale bar = 100µm. 2018-07-09100E-sv4.
Video 4:
Extension of hHES7-Achilles oscillations, related to Fig. 1. Timelapse imaging of human HES7-Achilles iPSC-derived PSM cells on days 2-4 in CLFBR medium. n=23 independent experiments. Scale bar = 100µm.
Video 5:
Conditions for automated cell tracking, related to Fig. 3. Sparse HES7-Achilles; pCAG-H2B-mCherry cells surrounded by wild type cells (no reporter) on days 2-3 of differentiation in CLFBR medium. n=16 independent experiments. Scale bar = 100µm.
Video 6:
Automated cell tracking and phase determination, related to Fig. 3. Overlay color-coded phases on automatically tracked HES7-Achilles; pCAG-H2B-mCherry cells over 15 hours on day 2 of differentiation in CLFBR medium. n=16 independent experiments.
Video 7:
Isolated cells, related to Fig. 3. hHES7-Achilles cells reseeded at low density on fibronectin-coated dishes. Left: DMSO (vehicle control), Center: Latrunculin A, Right: Latrunculin A + DAPT. n=5 independent experiments. Scale bar = 25 µm.
41586_2019_1885_MOESM11_ESM.mp4
Video 8: hHES7-Achilles; MESP2-mCherry reporter, related to Fig. 4. Timelapse imaging of human HES7-Achilles; MESP2-mCherry cells on days 2-5 of differentiation in CLFBR medium in control conditions, 25µM DAPT treatment, 2µM XAV treatment, and 250nM PD03 treatment. Note PD03-treated cells oscillate in opposite phase relative to all other conditions. n=10 independent experiments. Scale bar = 100µm.
Source data
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Diaz-Cuadros, M., Wagner, D.E., Budjan, C. et al. In vitro characterization of the human segmentation clock. Nature 580, 113–118 (2020). https://doi.org/10.1038/s41586-019-1885-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41586-019-1885-9
This article is cited by
-
Human neuronal maturation comes of age: cellular mechanisms and species differences
Nature Reviews Neuroscience (2024)
-
Ror homolog nhr-23 is essential for both developmental clock and circadian clock in C. elegans
Communications Biology (2024)
-
A patterned human neural tube model using microfluidic gradients
Nature (2024)
-
Cellular and molecular control of vertebrate somitogenesis
Nature Reviews Molecular Cell Biology (2024)
-
Morphogenesis beyond in vivo
Nature Reviews Physics (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.