Abstract
Peripheral artery disease (PAD) is a leading cause of cardiovascular morbidity and mortality; however, the extent to which genetic factors increase risk for PAD is largely unknown. Using electronic health record data, we performed a genome-wide association study in the Million Veteran Program testing ~32 million DNA sequence variants with PAD (31,307 cases and 211,753 controls) across veterans of European, African and Hispanic ancestry. The results were replicated in an independent sample of 5,117 PAD cases and 389,291 controls from the UK Biobank. We identified 19 PAD loci, 18 of which have not been previously reported. Eleven of the 19 loci were associated with disease in three vascular beds (coronary, cerebral, peripheral), including LDLR, LPL and LPA, suggesting that therapeutic modulation of low-density lipoprotein cholesterol, the lipoprotein lipase pathway or circulating lipoprotein(a) may be efficacious for multiple atherosclerotic disease phenotypes. Conversely, four of the variants appeared to be specific for PAD, including F5 p.R506Q, highlighting the pathogenic role of thrombosis in the peripheral vascular bed and providing genetic support for Factor Xa inhibition as a therapeutic strategy for PAD. Our results highlight mechanistic similarities and differences among coronary, cerebral and peripheral atherosclerosis and provide therapeutic insights.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
Data availability
The full summary level association data from the MVP transancestry PAD meta-analysis from this report are available through dbGAP, accession code phs001672.v2.p1. Additional data that support the findings of this study are available on request from the corresponding author (S.M.D.); these data are not publicly available due to US Government and Department of Veteran’s Affairs restrictions relating to participant privacy and consent. Data contributed by CARDIoGRAMplusC4D investigators are available online (http://www.CARDIOGRAMPLUSC4D.org/). Data on LAD have been contributed by the MEGASTROKE investigators and are available online (http://www.megastroke.org/). The genetic and phenotypic UK Biobank data are available upon application to the UK Biobank. Source data has been provided for Figs. 2 and 3 and Extended Data Figs. 4, 5 and 7. Additional data that were used to generate the figures in this study are available on request from the corresponding author (S.M.D.) or through dbGAP as listed above.
Code availability
Code to perform analyses in this manuscript are available from the authors upon request (D.K. and S.M.D.), or from the URLs associated with each software in the Methods section.
References
GBD 2016 Causes of Death Collaborators. Global, regional, and national age-sex specific mortality for 264 causes of death, 1980-2016: a systematic analysis for the Global Burden of Disease Study 2016. Lancet 390, 1151–1210 (2017).
Wahlgren, C. M. & Magnusson, P. K. Genetic influences on peripheral arterial disease in a twin population. Arterioscler Thromb. Vasc. Biol. 31, 678–682 (2011).
Murabito, J. M. et al. Association between chromosome 9p21 variants and the ankle-brachial index identified by a meta-analysis of 21 genome-wide association studies. Circ. Cardiovasc. Genet. 5, 100–112 (2012).
Matsukura, M. et al. Genome-wide association study of peripheral arterial disease in a Japanese population. PLoS ONE 10, e0139262 (2015).
Collins, R. What makes UK Biobank special? Lancet 379, 1173–1174 (2012).
Gaziano, J. M. et al. Million veteran program: A mega-biobank to study genetic influences on health and disease. J. Clin. Epidemiol. 70, 214–223 (2016).
Fan, J. et al. Billing code algorithms to identify cases of peripheral artery disease from administrative data. J. Am. Med Inf. Assoc. 20, e349–e354 (2013).
Kullo, I. J. et al. The ATXN2-SH2B3 locus is associated with peripheral arterial disease: an electronic medical record-based genome-wide association study. Front Genet. 5, 166 (2014).
Thorgeirsson, T. E. et al. A variant associated with nicotine dependence, lung cancer and peripheral arterial disease. Nature 452, 638–642 (2008).
Denny, J. C. et al. PheWAS: demonstrating the feasibility of a phenome-wide scan to discover gene-disease associations. Bioinformatics 26, 1205–1210 (2010).
Denny, J. C. et al. Systematic comparison of phenome-wide association study of electronic medical record data and genome-wide association study data. Nat. Biotechnol. 31, 1102–1110 (2013).
Voight, B. F. et al. Twelve type 2 diabetes susceptibility loci identified through large-scale association analysis. Nat. Genet. 42, 579–589 (2010).
Klarin, D., Emdin, C. A., Natarajan, P., Conrad, M. F. & Kathiresan, S. Genetic analysis of venous thromboembolism in UK biobank identifies the ZFPM2 locus and implicates obesity as a causal risk factor. Circ. Cardiovasc. Genet. 10, e001643 (2017).
Teslovich, T. M. et al. Biological, clinical and population relevance of 95 loci for blood lipids. Nature 466, 707–713 (2010).
de Bakker, P. I. et al. A high-resolution HLA and SNP haplotype map for disease association studies in the extended human MHC. Nat. Genet. 38, 1166–1172 (2006).
Conte, M. S. et al. Society for Vascular Surgery practice guidelines for atherosclerotic occlusive disease of the lower extremities: management of asymptomatic disease and claudication. J. Vasc. Surg. 61, 2s–41s (2015).
Staley, J. R. et al. PhenoScanner: a database of human genotype-phenotype associations. Bioinformatics 32, 3207–3209 (2016).
GTEx Consortium. Genetic effects on gene expression across human tissues. Nature 550, 204–213 (2017).
Di Angelantonio, E. et al. Efficiency and safety of varying the frequency of whole blood donation (INTERVAL): a randomised trial of 45 000 donors. Lancet 390, 2360–2371 (2017).
Sun, B. B. et al. Genomic atlas of the human plasma proteome. Nature 558, 73–79 (2018).
Gusev, A. et al. Integrative approaches for large-scale transcriptome-wide association studies. Nat. Genet. 48, 245–252 (2016).
Anttila, V. et al. Analysis of shared heritability in common disorders of the brain. Science 360, eaap8757 (2018).
Bulik-Sullivan, B. et al. An atlas of genetic correlations across human diseases and traits. Nat. Genet. 47, 1236–1241 (2015).
CARDIoGRAMplusC4D Consortium. A comprehensive 1000 Genomes-based genome-wide association meta-analysis of coronary artery disease. Nat. Genet. 47, 1121–1130 (2015).
Malik, R. et al. Multiancestry genome-wide association study of 520,000 subjects identifies 32 loci associated with stroke and stroke subtypes. Nat. Genet. 50, 524–537 (2018).
Sibon, I. et al. COL4A1 mutation in Axenfeld–Rieger anomaly with leukoencephalopathy and stroke. Ann. Neurol. 62, 177–184 (2007).
Greengard, J. S., Eichinger, S., Griffin, J. H. & Bauer, K. A. Brief report: variability of thrombosis among homozygous siblings with resistance to activated protein C due to an Arg—>Gln mutation in the gene for factor V. N. Engl. J. Med. 331, 1559–1562 (1994).
Bertina, R. M. et al. Mutation in blood coagulation factor V associated with resistance to activated protein C. Nature 369, 64–67 (1994).
Holst, A. G., Jensen, G. & Prescott, E. Risk factors for venous thromboembolism: results from the Copenhagen City Heart Study. Circulation 121, 1896–1903 (2010).
Cheng, Y. J. et al. Current and former smoking and risk for venous thromboembolism: a systematic review and meta-analysis. PLoS Med. 10, e1001515 (2013).
Willey, J. et al. Epidemiology of lower extremity peripheral artery disease in veterans. J. Vasc. Surg. 68, 527–535.e5 (2018).
Fowkes, F. G. et al. Comparison of global estimates of prevalence and risk factors for peripheral artery disease in 2000 and 2010: a systematic review and analysis. Lancet 382, 1329–1340 (2013).
Myocardial Infarction Genetics and CARDIoGRAM Exome Consortia Investigators. Coding Variation in ANGPTL4, LPL, and SVEP1 and the Risk of Coronary Disease. N. Engl. J. Med. 374, 1134–1144 (2016).
Musunuru, K. et al. Exome sequencing, ANGPTL3 mutations, and familial combined hypolipidemia. N. Engl. J. Med. 363, 2220–2227 (2010).
Dewey, F. E. et al. Genetic and pharmacologic inactivation of ANGPTL3 and cardiovascular disease. N. Engl. J. Med. 377, 211–221 (2017).
Emdin, C. A. et al. Phenotypic characterization of genetically lowered human lipoprotein(a) levels. J. Am. Coll. Cardiol. 68, 2761–2772 (2016).
Swanberg, M. et al. MHC2TA is associated with differential MHC molecule expression and susceptibility to rheumatoid arthritis, multiple sclerosis and myocardial infarction. Nat. Genet. 37, 486–494 (2005).
Anand, S. S. et al. Rivaroxaban with or without aspirin in patients with stable peripheral or carotid artery disease: an international, randomised, double-blind, placebo-controlled trial. Lancet 391, 219–229 (2017).
The 1000 Genomes Project Consortium. A global reference for human genetic variation. Nature 526, 68–74 (2015).
Loh, P. R., Palamara, P. F. & Price, A. L. Fast and accurate long-range phasing in a UK Biobank cohort. Nat. Genet. 48, 811–816 (2016).
Howie, B., Fuchsberger, C., Stephens, M., Marchini, J. & Abecasis, G. R. Fast and accurate genotype imputation in genome-wide association studies through pre-phasing. Nat. Genet. 44, 955–959 (2012).
Price, A. L. et al. Principal components analysis corrects for stratification in genome-wide association studies. Nat. Genet. 38, 904–909 (2006).
Alexander, D. H., Novembre, J. & Lange, K. Fast model-based estimation of ancestry in unrelated individuals. Genome Res. 19, 1655–1664 (2009).
Klarin, D. et al. Genetics of blood lipids among ~300,000 multi-ethnic participants of the Million Veteran Program. Nat. Genet. 50, 1514–1523 (2018).
Manichaikul, A. et al. Robust relationship inference in genome-wide association studies. Bioinformatics 26, 2867–2873 (2010).
Winkler, T. W. et al. Quality control and conduct of genome-wide association meta-analyses. Nat. Protoc. 9, 1192–1212 (2014).
Hyde, C. L. et al. Identification of 15 genetic loci associated with risk of major depression in individuals of European descent. Nat. Genet. 48, 1031–1036 (2016).
Bycroft, C. et al. The UK Biobank resource with deep phenotyping and genomic data. Nature 562, 203–209 (2018).
Bellenguez, C., Strange, A., Freeman, C., Donnelly, P. & Spencer, C. C. A robust clustering algorithm for identifying problematic samples in genome-wide association studies. Bioinformatics 28, 134–135 (2012).
Arya, S. et al. Race and socioeconomic status independently affect risk of major amputation in peripheral artery disease. J. Am. Heart Assoc. 7, e007425 (2018).
Song, R. J. et al. Abstract 18809: Development of an electronic health record-based algorithm for smoking status using the Million Veteran Program (MVP) cohort survey response. Circulation 134 A18809 (2016).
Carroll, R. J., Bastarache, L. & Denny, J. C. R PheWAS: data analysis and plotting tools for phenome-wide association studies in the R environment. Bioinformatics 30, 2375–2376 (2014).
Zhou, X., Carbonetto, P. & Stephens, M. Polygenic modeling with bayesian sparse linear mixed models. PLoS Genet. 9, e1003264 (2013).
Willer, C. J., Li, Y. & Abecasis, G. R. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics 26, 2190–2191 (2010).
Alba, P. R. et al. Ankle brachial index extraction system. In AMIA Annu. Symp. Proc. https://symposium2018.zerista.com/event/member?item_id=8296486 2018.
Cornia, R., Patterson, O. V., Ginter, T. & DuVall, S. L. Rapid NLP development with Leo. In AMIA Annual Symposium Proc eedings 1356 (2014).
Ferrucci, D. & Lally, A. UIMA: an architectural approach to unstructured information processing in the corporate research environment. Nat. Lang. Eng. 10, 327–348 (2004).
Acknowledgements
This research is based on data from the MVP, Office of Research and Development, Veterans Health Administration and was supported by award no. MVP000. This publication does not represent the views of the Department of Veterans Affairs, the US Food and Drug Administration, or the US Government. This research was also supported by funding from: the Department of Veterans Affairs awards nos. I01-BX03340 (K.C. and P.W.F.W.), I01-BX003362 (P.S.T. and K.M.C) and IK2-CX001780 (S.M.D.) and the VA Informatics and Computing Infrastructure (VINCI) VA HSR RES 130457 (P.A., O.V.P. and S.L.D.); the National Institutes of Health grants nos. R01-HL131977 (J.A.B.), R03-AG050930 (S.A.), R01-HL138306 (J.Chen), R01-HL127564 (S.K.) and K08-HL140203 (P.N.); and the American Heart Association grant no. 18SFRN33960373 (J.A.B. and M.S.F.), 17IFUNP33840012 (K.A.) and 15MCPRP25580005 (S.A.). Data on CAD have been contributed by the CARDIoGRAMplusC4D investigators. Data on large artery stroke have been contributed by the MEGASTROKE investigators. The MEGASTROKE project received funding from sources specified at http://www.megastroke.org/acknowledgements.html.
Author information
Authors and Affiliations
Consortia
Contributions
D.K., J.L., T.L.A., K.A., M.C., M.S.F., J.A.B., J.M.G., J. Concato, D.J.R., K.C., K.-M.C., P.W.F.W., C.J.O., S.K., P.S.T. and S.M.D were responsible for the concept and design. The acquisition, analysis or interpretation of data were performed by D.K., J.L., K.A., M.C., T.L.A., J.H., K.M.L., Q.S., J.E.H., P.N., S.A., A.S., Y.V.S., M.V., M.S.F., L.W., J. Chen, D.S., J.S.L., D.R.M., P.R., P.R.A., O.V.P., S.L.D., W.E.B., J.A.B., J. Concato, J.M.G., D.J.R., K.C., K.-M.C., P.W.F.W., C.J.O., S.K., P.S.T. and S.M.D. The authors D.K., P.S.T. and S.M.D. drafted the manuscript. The critical revision of the manuscript for important intellectual content was carried out by D.K., T.L.A., Y.V.S., K.C., P.N., D.J.R., K.C., K.-M.C., C.J.O., J.A.B., M.S.F., O.V.P., P.W.W., S.K., P.S.T. and S.M.D. The authors D.K., P.N., Y.V.S., M.S.F., J. Chen, J.A.B., J. Concato, J.M.G., D.J.R., K.C., K.-M.C., C.J.O., P.W.F.W., S.K., P.S.T. and S.M.D. provided administrative, technical, or material support.
Corresponding author
Ethics declarations
Competing interests
J.A.B. reports consulting with AstraZeneca, Bristol Myers Squibb, Amgen, Merck, Sanofi, Antidote Pharmaceutical and Boehringer Ingelheim. He serves on the DSMC for Bayer and Novartis. S.L.D. has received research grant support from the following for-profit companies through the University of Utah or the Western Institute for Biomedical Research (VA Salt Lake City’s affiliated non-profit): AbbVie Inc., Anolinx LLC, Astellas Pharma Inc., AstraZeneca Pharmaceuticals LP, Boehringer Ingelheim International GmbH, Celgene Corporation, Eli Lilly and Company, Genentech Inc., Genomic Health, Inc., Gilead Sciences Inc., GlaxoSmithKline PLC, Innocrin Pharmaceuticals Inc., Janssen Pharmaceuticals, Inc., Kantar Health, Myriad Genetic Laboratories, Inc., Novartis International AG and PAREXEL International Corporation. S.M.D. receives research support to his institution from CytoVAS and RenalytixAI. S.K. is a founder of Maze Therapeutics, Verve Therapeutics and San Therapeutics. He holds equity in Catabasis and San Therapeutics. He is a member of the scientific advisory boards for Regeneron Genetics Center and Corvidia Therapeutics; served as a consultant for Acceleron, Eli Lilly, Novartis, Merck, NovoNordisk, Novo Ventures, Ionis, Alnylam, Aegerion, Huag Partners, Noble Insights, Leerink Partners, Bayer Healthcare, Illumina, Color Genomics, MedGenome, Quest and Medscape; and reports patents related to a method of identifying and treating a person having a predisposition to or afflicted with cardiometabolic disease (20180010185) and a genetics risk predictor (20190017119). O.V.P. has received research grants from the following for-profit organizations through the University of Utah or Western Institute for Biomedical Research: Anolinx LLC, AstraZeneca Pharmaceuticals LP, Genentech Inc., Genomic Health, Inc., Gilead Sciences Inc., Janssen Pharmaceuticals, Inc., Novartis International AG and PAREXEL International Corporation.
Additional information
Peer review information: Michael Basson was the primary editor on this article and managed its editorial process and peer review in collaboration with the rest of the editorial team.
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Distribution of minimum ankle–brachial index values in the MVP.
Histogram of minimum mABI values extracted from the electronic health record for 17,861 participants of the MVP. These values, restricted to those with an minimum ABI of <1.4, were used for the subsequent mABI GWAS.
Extended Data Fig. 2 Quantile-quantile plot for the discovery trans-ethnic PAD GWAS in MVP.
The expected logistic regression association P values versus the observed distribution of P values for PAD association (N = 31,307 PAD cases and 211,753 controls) are displayed. Quantile–quantile plots were inspected for ancestry-specific analyses and genomic control values were <1.20 for each racial group (data not shown). No systemic inflation was observed (λGC = 1.05). All P values were two-sided.
Extended Data Fig. 3 Manhattan plot for the PAD GWAS.
Plot of –log10(P) for association of imputed variants by chromosomal position for all autosomal polymorphisms analyzed in the PAD GWAS (N = 36,424 PAD cases and 601,044 controls). The genes nearest to the top associated variants are displayed. Genes highlighted in red represent novel PAD loci (18). Genes for variants that are outside the transcript boundary of a protein-coding gene are shown with nearest candidate gene in parentheses (for example, (LDLR)). Logistic regression two-sided P values are displayed.
Extended Data Fig. 4 TCF7L2 mediates its effect on PAD via T2D.
a, Forest plot depicting the replication of the known TCF7L2/rs7903146-T2D association signal12 in MVP for both white and black participants. b, The same variant is also associated with PAD risk in whites and blacks in MVP. However, when controlling for T2D status in the regression model, c, the association signal is dramatically reduced, suggesting that TCF7L2 PAD risk is mediated through its effect on T2D. Logistic regression two-sided values of P are displayed. Gray boxes reflect the inverse-variance weight for each ancestry.
Extended Data Fig. 5 Forest plot for association of the CHRNA3 locus and peripheral artery disease risk stratified by smoking status.
When stratifying European MVP participants by smoking status (ever smokers versus never smokers), nearly all the association signal resides within the ever smoker group. Previous reports of variation at the CHRNA3 locus demonstrate that carriers of the PAD risk allele have a reduced likelihood of cigarette smoking cessation9. This suggests that the PAD-CHRNA3 association is driven by a greater burden of tobacco exposure in those who carry the nicotine dependence/PAD risk allele. Logistic regression two-sided values of P are displayed. Gray boxes reflect the inverse-variance weight for each subgroup.
Extended Data Fig. 6 Peripheral artery disease risk loci and known causal risk factors.
Peripheral artery disease risk loci identified in this GWAS analysis are depicted along with the plausible relationship to the underling causal risk factor. Loci names are based on the nearest genes; however, the causal gene(s) remains unclear for some associated loci and, as such, the resultant annotation may prove incorrect in some cases.
Extended Data Fig. 7 Peripheral artery disease risk variants and association with LAS and CAD.
For the 19 PAD risk variants identified in our study, logistic regression Z-scores of association (aligned to the PAD risk allele) were obtained from MVP (PAD, N = 31,307 PAD cases and 211,753 controls) and publicly available summary statistics for large artery stroke (MVP+MEGASTROKE consortium25, N = 7,393 LAS cases and 628,737 controls) and coronary artery disease (MVP+CARDIoGRAMplusC4D consortium24, N = 111,216 CAD cases and 248,081 controls). A positive Z-score (red) indicates a positive association between the PAD risk allele and the disease, while a negative Z-score (blue) indicates an inverse association. Boxes are outlined in cyan if the variant is uniquely associated with PAD (two-sided logistic regression PPAD < 5 × 10−8, PCAD and PLAS > 0.05).
Extended Data Fig. 8 Peripheral artery disease risk variants and mechanistic overlap with LAS and CAD.
Venn diagram of each of the 19 PAD risk loci in a based on their association with PAD (N = 31,307 PAD cases and 211,753 controls; two-sided PPAD < 5 × 10−8), CAD (N = 111,216 CAD cases and 248,081 controls; two-sided P < 0.05) and LAS (N = 7,393 LAS cases and 628,737 controls; two-sided P < 0.05) using logistic regression. Each locus is depicted along with the plausible relationship to the underling causal risk factor separately by color. Loci names are based on the nearest genes; however, the causal gene(s) remains unclear for some associated loci and as such, the resultant annotation may prove incorrect in some cases.
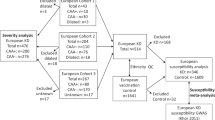
Extended Data Fig. 9 Overall study design.
The primary analysis consisted of a genome-wide association study to identify novel PAD risk variants. Secondary analyses involved a genome-wide association study of minimum ABI, a closer examination the 19 PAD risk variants through PheWAS, a candidate causal gene analysis using eQTL/pQTL/TWAS data, a PAD analysis accounting for CAD/LAS status and a focused Factor V Leiden analysis.
Extended Data Fig. 10 Natural language processing for index extraction.
Examples of semi-structured text that contains targeted indices for extraction using natural language processing (NLP). TBI, toe–brachial index; PT, posterior tibial artery; AT, anterior tibial artery.
Supplementary information
Supplementary Information
Supplementary Tables 1, 3, 13, 15; MVP and MEGASTROKE investigators
Supplementary Tables
Supplementary Tables 2, 4, 5–12, 14, 16–26
Source data
Source Data Fig. 2
Raw Z Scores used for HeatMap Creation
Source Data Fig. 3
Raw Association Statistics for Forest Plot Creation
Source Data Extended Data Fig. 4
Raw Association Statistics for Forest Plot Creation
Source Data Extended Data Fig. 5
Raw Association Statistics for Forest Plot Creation
Source Data Extended Data Fig. 7
Raw Z Scores used for HeatMap Creation
Rights and permissions
About this article
Cite this article
Klarin, D., Lynch, J., Aragam, K. et al. Genome-wide association study of peripheral artery disease in the Million Veteran Program. Nat Med 25, 1274–1279 (2019). https://doi.org/10.1038/s41591-019-0492-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41591-019-0492-5
This article is cited by
-
A linear weighted combination of polygenic scores for a broad range of traits improves prediction of coronary heart disease
European Journal of Human Genetics (2024)
-
A cost-effective sequencing method for genetic studies combining high-depth whole exome and low-depth whole genome
npj Genomic Medicine (2024)
-
Prevalence and demographic variation of cardiovascular, renal, metabolic, and mental health conditions in 12 million english primary care records
BMC Medical Informatics and Decision Making (2023)
-
Cold-induced vasodilation response in a Japanese cohort: insights from cold-water immersion and genome-wide association studies
Journal of Physiological Anthropology (2023)
-
A multi-ancestry polygenic risk score improves risk prediction for coronary artery disease
Nature Medicine (2023)