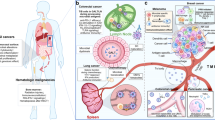
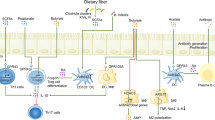
Abstract
Microorganisms within the gut and other niches may contribute to carcinogenesis, as well as shaping cancer immunosurveillance and response to immunotherapy. Our understanding of the complex relationship between different host-intrinsic microorganisms, as well as the multifaceted mechanisms by which they influence health and disease, has grown tremendously—hastening development of novel therapeutic strategies that target the microbiota to improve treatment outcomes in cancer. Accordingly, the evaluation of a patient’s microbial composition and function and its subsequent targeted modulation represent key elements of future multidisciplinary and precision-medicine approaches. In this Review, we outline the current state of research toward harnessing the microbiome to better prevent and treat cancer.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Dodd, M. S. et al. Evidence for early life in Earth’s oldest hydrothermal vent precipitates. Nature 543, 60–64 (2017).
Dismukes, G. C. et al. The origin of atmospheric oxygen on Earth: the innovation of oxygenic photosynthesis. Proc. Natl Acad. Sci. USA 98, 2170–2175 (2001).
Fullerton, K. M. et al. Effect of tectonic processes on biosphere–geosphere feedbacks across a convergent margin. Nat. Geosci. 14, 301–306 (2021).
Friedrich, M. J. Genomes of microbes inhabiting the body offer clues to human health and disease. J. Am. Med. Assoc. 309, 1447–1449 (2013).
Neish, A. S. Microbes in gastrointestinal health and disease. Gastroenterology 136, 65–80 (2009).
Young, V. B. The role of the microbiome in human health and disease: an introduction for clinicians. Br. Med. J. 356, j831 (2017).
Barrett, K. E. & Wu, G. D. Influence of the microbiota on host physiology—moving beyond the gut. J. Physiol. 595, 433–435 (2017).
Dominguez-Bello, M. G., Godoy-Vitorino, F., Knight, R. & Blaser, M. J. Role of the microbiome in human development. Gut 68, 1108–1114 (2019).
Reck, M. et al. Pembrolizumab versus chemotherapy for PD-L1-positive non-small-cell lung cancer. N. Engl. J. Med. 375, 1823–1833 (2016).
Robert, C. et al. Nivolumab in previously untreated melanoma without BRAF mutation. N. Engl. J. Med. 372, 320–330 (2015).
Weber, J. S. et al. Nivolumab versus chemotherapy in patients with advanced melanoma who progressed after anti-CTLA-4 treatment (CheckMate 037): a randomised, controlled, open-label, phase 3 trial. Lancet Oncol. 16, 375–384 (2015).
Rini, B. I. et al. Pembrolizumab plus axitinib versus sunitinib for advanced renal cell carcinoma. N. Engl. J. Med. 380, 1116–1127 (2019).
Overman, M. J. et al. Durable clinical benefit with nivolumab plus ipilimumab in DNA mismatch repair-deficient/microsatellite instability-high metastatic colorectal cancer. J. Clin. Oncol. 36, 773–779 (2018).
Sharma, P., Hu-Lieskovan, S., Wargo, J. A. & Ribas, A. Primary, adaptive, and acquired resistance to cancer immunotherapy. Cell 168, 707–723 (2017).
Gide, T. N., Wilmott, J. S., Scolyer, R. A. & Long, G. V. Primary and acquired resistance to immune checkpoint inhibitors in metastatic melanoma. Clin. Cancer Res. 24, 1260–1270 (2018).
Brahmer, J. R. et al. The Society for Immunotherapy of Cancer consensus statement on immunotherapy for the treatment of non-small cell lung cancer (NSCLC). J. Immunother. Cancer 6, 75 (2018).
Gandhi, L. et al. Pembrolizumab plus chemotherapy in metastatic non-small-cell lung cancer. N. Engl. J. Med. 378, 2078–2092 (2018).
Fehrenbacher, L. et al. Atezolizumab versus docetaxel for patients with previously treated non-small-cell lung cancer (POPLAR): a multicentre, open-label, phase 2 randomised controlled trial. Lancet 387, 1837–1846 (2016).
Morad, G., Helmink, B. A., Sharma, P. & Wargo, J. A. Hallmarks of response, resistance, and toxicity to immune checkpoint blockade. Cell 184, 5309–5337 (2021).
Le, D. T. et al. PD-1 blockade in tumors with mismatch-repair deficiency. N. Engl. J. Med. 372, 2509–2520 (2015).
Ayers, M. et al. IFN-gamma-related mRNA profile predicts clinical response to PD-1 blockade. J. Clin. Investig. 127, 2930–2940 (2017).
Liu, D. et al. Integrative molecular and clinical modeling of clinical outcomes to PD-1 blockade in patients with metastatic melanoma. Nat. Med. 25, 1916–1927 (2019).
Bodor, J. N., Boumber, Y. & Borghaei, H. Biomarkers for immune checkpoint inhibition in non-small-cell lung cancer (NSCLC). Cancer 126, 260–270 (2020).
Dong, Z. Y. et al. Potential predictive value of TP53 and KRAS mutation status for response to PD-1 blockade immunotherapy in lung adenocarcinoma. Clin. Cancer Res. 23, 3012–3024 (2017).
Gopalakrishnan, V. et al. Gut microbiome modulates response to anti-PD-1 immunotherapy in melanoma patients. Science 359, 97–103 (2018).
Routy, B. et al. Gut microbiome influences efficacy of PD-1-based immunotherapy against epithelial tumors. Science 359, 91–97 (2018).
Yonekura, S. et al. Cancer induces a stress ileopathy depending on B-adrenergic receptors and promoting dysbiosis that contribute to carcinogenesis. Cancer Discov. 12, 1128–1151 (2022).
Spencer, C. N. et al. Dietary fiber and probiotics influence the gut microbiome and melanoma immunotherapy response. Science 374, 1632–1640 (2021).
Davar, D. et al. Fecal microbiota transplant overcomes resistance to anti-PD-1 therapy in melanoma patients. Science 371, 595–602 (2021).
Baruch, E. N. et al. Fecal microbiota transplant promotes response in immunotherapy-refractory melanoma patients. Science 371, 602–609 (2021).
Wang, D. D. et al. The gut microbiome modulates the protective association between a Mediterranean diet and cardiometabolic disease risk. Nat. Med. 27, 333–343 (2021).
Andrews, M. C. et al. Gut microbiota signatures are associated with toxicity to combined CTLA-4 and PD-1 blockade. Nat. Med. 27, 1432–1441 (2021).
Blake, S. J. et al. The immunotoxicity, but not anti-tumor efficacy, of anti-CD40 and anti-CD137 immunotherapies is dependent on the gut microbiota. Cell Rep. Med 2, 100464 (2021).
Sepich-Poore, G. D. et al. The microbiome and human cancer. Science 371, eabc4552 (2021).
Cogdill, A. P., Gaudreau, P. O., Arora, R., Gopalakrishnan, V. & Wargo, J. A. The impact of intratumoral and gastrointestinal microbiota on systemic cancer therapy. Trends Immunol. 39, 900–920 (2018).
Heymann, C. J. F., Bard, J. M., Heymann, M. F., Heymann, D. & Bobin-Dubigeon, C. The intratumoral microbiome: characterization methods and functional impact. Cancer Lett. 522, 63–79 (2021).
Nejman, D. et al. The human tumor microbiome is composed of tumor type-specific intracellular bacteria. Science 368, 973–980 (2020).
Liou, J.-M. et al. Screening and eradication of Helicobacter pylori for gastric cancer prevention: the Taipei global consensus. Gut 69, 2093–2112 (2020).
Cao, Y. EBV-based cancer prevention and therapy in nasopharyngeal carcinoma. NPJ Precis. Oncol. 1, 10 (2017).
Poore, G. D. et al. Microbiome analyses of blood and tissues suggest cancer diagnostic approach. Nature 579, 567–574 (2020).
Lei, J. et al. HPV vaccination and the risk of invasive cervical cancer. N. Engl. J. Med. 383, 1340–1348 (2020).
Chang, M. H. et al. Prevention of hepatocellular carcinoma by universal vaccination against hepatitis B virus: the effect and problems. Clin. Cancer Res. 11, 7953–7957 (2005).
Yu, T. et al. Fusobacterium nucleatum promotes chemoresistance to colorectal cancer by modulating autophagy. Cell 170, 548–563 (2017).
Knippel, R. J., Drewes, J. L. & Sears, C. L. The cancer microbiome: recent highlights and knowledge gaps. Cancer Discov. 11, 2378–2395 (2021).
Lynch, S. V. & Pedersen, O. The human intestinal microbiome in health and disease. N. Engl. J. Med. 375, 2369–2379 (2016).
Helmink, B. A., Khan, M. A. W., Hermann, A., Gopalakrishnan, V. & Wargo, J. A. The microbiome, cancer, and cancer therapy. Nat. Med. 25, 377–388 (2019).
Hanahan, D. Hallmarks of cancer: new dimensions. Cancer Discov. 12, 31–46 (2022).
Matson, V. et al. The commensal microbiome is associated with anti-PD-1 efficacy in metastatic melanoma patients. Science 359, 104–108 (2018).
Sivan, A. et al. Commensal Bifidobacterium promotes antitumor immunity and facilitates anti-PD-L1 efficacy. Science 350, 1084–1089 (2015).
Vetizou, M. et al. Anticancer immunotherapy by CTLA-4 blockade relies on the gut microbiota. Science 350, 1079–1084 (2015).
Derosa, L. et al. Microbiota-centered interventions: the next breakthrough in immuno-oncology? Cancer Discov. 11, 2396–2412 (2021).
Roberti, M. P. et al. Chemotherapy-induced ileal crypt apoptosis and the ileal microbiome shape immunosurveillance and prognosis of proximal colon cancer. Nat. Med. 26, 919–931 (2020).
Goubet, A. G. et al. Multifaceted modes of action of the anticancer probiotic Enterococcus hirae. Cell Death Differ. 28, 2276–2295 (2021).
Plovier, H. et al. A purified membrane protein from Akkermansia muciniphila or the pasteurized bacterium improves metabolism in obese and diabetic mice. Nat. Med. 23, 107–113 (2017).
Griffin, M. E. et al. Enterococcus peptidoglycan remodeling promotes checkpoint inhibitor cancer immunotherapy. Science 373, 1040–1046 (2021).
Yoon, H. S. et al. Akkermansia muciniphila secretes a glucagon-like peptide-1-inducing protein that improves glucose homeostasis and ameliorates metabolic disease in mice. Nat. Microbiol. 6, 563–573 (2021).
Grajeda-Iglesias, C. et al. Oral administration of Akkermansia muciniphila elevates systemic antiaging and anticancer metabolites. Aging 13, 6375–6405 (2021).
Bessell, C. A. et al. Commensal bacteria stimulate antitumor responses via T cell cross-reactivity. JCI Insight 5, e135597 (2020).
Fluckiger, A. et al. Cross-reactivity between tumor MHC class I-restricted antigens and an enterococcal bacteriophage. Science 369, 936–942 (2020).
He, Y. et al. Gut microbial metabolites facilitate anticancer therapy efficacy by modulating cytotoxic CD8+ T cell immunity. Cell Metab. 33, 988–1000 (2021).
Bultman, S. J. Molecular pathways: gene–environment interactions regulating dietary fiber induction of proliferation and apoptosis via butyrate for cancer prevention. Clin. Cancer Res. 20, 799–803 (2014).
Belcheva, A. et al. Gut microbial metabolism drives transformation of MSH2-deficient colon epithelial cells. Cell 158, 288–299 (2014).
Kim, K. et al. Propionate of a microbiota metabolite induces cell apoptosis and cell cycle arrest in lung cancer. Mol. Med. Rep. 20, 1569–1574 (2019).
Bindels, L. B. et al. Gut microbiota-derived propionate reduces cancer cell proliferation in the liver. Br. J. Cancer 107, 1337–1344 (2012).
Geller, L. T. et al. Potential role of intratumor bacteria in mediating tumor resistance to the chemotherapeutic drug gemcitabine. Science 357, 1156–1160 (2017).
Yan, A. et al. Transformation of the anticancer drug doxorubicin in the human gut microbiome. ACS Infect. Dis. 4, 68–76 (2018).
Westman, E. L. et al. Bacterial inactivation of the anticancer drug doxorubicin. Chem. Biol. 19, 1255–1264 (2012).
Ruf, B. et al. Activating mucosal-associated Invariant T cells induces a broad antitumor response. Cancer Immunol. Res. 9, 1024–1034 (2021).
Legoux, F. et al. Microbial metabolites control the thymic development of mucosal-associated invariant T cells. Science 366, 494–499 (2019).
Mager, L. F. et al. Microbiome-derived inosine modulates response to checkpoint inhibitor immunotherapy. Science 369, 1481–1489 (2020).
Pernigoni, N. et al. Commensal bacteria promote endocrine resistance in prostate cancer through androgen biosynthesis. Science 374, 216–224 (2021).
Fuhrman, B. J. et al. Associations of the fecal microbiome with urinary estrogens and estrogen metabolites in postmenopausal women. J. Clin. Endocrinol. Metab. 99, 4632–4640 (2014).
Flores, R. et al. Fecal microbial determinants of fecal and systemic estrogens and estrogen metabolites: a cross-sectional study. J. Transl. Med. 10, 253 (2012).
Bauer, H., Horowitz, R. E., Levenson, S. M. & Popper, H. The response of the lymphatic tissue to the microbial flora. Studies on germ-free mice. Am. J. Pathol. 42, 471–483 (1963).
Pabst, O. et al. Adaptation of solitary intestinal lymphoid tissue in response to microbiota and chemokine receptor CCR7 signaling. J. Immunol. 177, 6824–6832 (2006).
Lathrop, S. K. et al. Peripheral education of the immune system by colonic commensal microbiota. Nature 478, 250–254 (2011).
Kamada, N., Seo, S. U., Chen, G. Y. & Nunez, G. Role of the gut microbiota in immunity and inflammatory disease. Nat. Rev. Immunol. 13, 321–335 (2013).
Abt, M. C. et al. Commensal bacteria calibrate the activation threshold of innate antiviral immunity. Immunity 37, 158–170 (2012).
Huhta, H. et al. Toll-like receptors 1, 2, 4 and 6 in esophageal epithelium, Barrett’s esophagus, dysplasia and adenocarcinoma. Oncotarget 7, 23658–23667 (2016).
Ge, Y. et al. Gut microbiota influence tumor development and alter interactions with the human immune system. J. Exp. Clin. Cancer Res. 40, 42 (2021).
Lam, K. C. et al. Microbiota triggers STING-type I IFN-dependent monocyte reprogramming of the tumor microenvironment. Cell 184, 5338–5356 (2021).
Chen, G. Y., Shaw, M. H., Redondo, G. & Nunez, G. The innate immune receptor Nod1 protects the intestine from inflammation-induced tumorigenesis. Cancer Res. 68, 10060–10067 (2008).
Bouskra, D. et al. Lymphoid tissue genesis induced by commensals through NOD1 regulates intestinal homeostasis. Nature 456, 507–510 (2008).
Mazmanian, S. K., Liu, C. H., Tzianabos, A. O. & Kasper, D. L. An immunomodulatory molecule of symbiotic bacteria directs maturation of the host immune system. Cell 122, 107–118 (2005).
Viaud, S. et al. The intestinal microbiota modulates the anticancer immune effects of cyclophosphamide. Science 342, 971–976 (2013).
Iida, N. et al. Commensal bacteria control cancer response to therapy by modulating the tumor microenvironment. Science 342, 967–970 (2013).
Derosa, L. et al. Impact of antibiotics on outcome in patients with metastatic renal cell carcinoma treated with immune checkpoint inhibitors. J. Clin. Oncol. 35, 462–462 (2017).
Huemer, F. et al. Association between antibiotics use and outcome in patients with NSCLC treated with immunotherapeutics. Ann. Oncol. 30, 652–653 (2019).
Khan, U. et al. Impact of use of antibiotics on response to immune checkpoint inhibitors and tumor microenvironment. Am. J. Clin. Oncol. 44, 247–253 (2021).
Serpas, V. et al. Impact of antibiotic exposure on the efficacy of immune checkpoint blockade in MSI-H metastatic CRC. J. Clin. Oncol. 38, 161–161 (2020).
Lee, K. A. et al. Cross-cohort gut microbiome associations with immune checkpoint inhibitor response in advanced melanoma. Nat. Med. https://doi.org/10.1038/s41591-022-01695-5 (2022).
Derosa, L. et al. Intestinal Akkermansia muciniphila predicts clinical response to PD-1 blockade in patients with advanced non-small-cell lung cancer. Nat. Med. 28, 315–324 (2022).
Hakozaki, T. et al. The gut microbiome associates with immune checkpoint inhibition outcomes in patients with advanced non-small-cell lung cancer. Cancer Immunol. Res. 8, 1243–1250 (2020).
Cascone, T. et al. Neoadjuvant nivolumab or nivolumab plus ipilimumab in operable non-small cell lung cancer: the phase 2 randomized NEOSTAR trial. Nat. Med. 27, 504–514 (2021).
Jin, Y. et al. The diversity of gut microbiome is associated with favorable responses to anti-programmed death 1 immunotherapy in Chinese patients with NSCLC. J. Thorac. Oncol. 14, 1378–1389 (2019).
Derosa, L. et al. Gut bacteria composition drives primary resistance to cancer immunotherapy in renal cell carcinoma patients. Eur. Urol. 78, 195–206 (2020).
Salgia, N. J. et al. Stool microbiome profiling of patients with metastatic renal cell carcinoma receiving anti-PD-1 immune checkpoint inhibitors. Eur. Urol. 78, 498–502 (2020).
Mao, J. et al. Gut microbiome is associated with the clinical response to anti-PD-1 based immunotherapy in hepatobiliary cancers. J. Immunother. Cancer 9, e003334 (2021).
Peng, Z. et al. The gut microbiome is associated with clinical response to anti-PD-1/PD-L1 immunotherapy in gastrointestinal cancer. Cancer Immunol. Res. 8, 1251–1261 (2020).
Zheng, Y. et al. Gut microbiome affects the response to anti-PD-1 immunotherapy in patients with hepatocellular carcinoma. J. Immunother. Cancer 7, 193 (2019).
Gharaibeh, R. Z. & Jobin, C. Microbiota and cancer immunotherapy: in search of microbial signals. Gut 68, 385–388 (2019).
Mitra, A. et al. Microbial diversity and composition is associated with patient-reported toxicity during chemoradiation therapy for cervical cancer. Int. J. Radiat. Oncol. Biol. Phys. 107, 163–171 (2020).
Wang, A. et al. Gut microbial dysbiosis may predict diarrhea and fatigue in patients undergoing pelvic cancer radiotherapy: a pilot study. PLoS ONE 10, e0126312 (2015).
Biagi, E. et al. Early gut microbiota signature of aGvHD in children given allogeneic hematopoietic cell transplantation for hematological disorders. BMC Med. Genomics 12, 49 (2019).
Biagi, E. et al. Gut microbiota trajectory in pediatric patients undergoing hematopoietic SCT. Bone Marrow Transplant. 50, 992–998 (2015).
Jenq, R. R. et al. Intestinal blautia is associated with reduced death from graft-versus-host disease. Biol. Blood Marrow Transplant. 21, 1373–1383 (2015).
Holler, E. et al. Metagenomic analysis of the stool microbiome in patients receiving allogeneic stem cell transplantation: loss of diversity is associated with use of systemic antibiotics and more pronounced in gastrointestinal graft-versus-host disease. Biol. Blood Marrow Transplant. 20, 640–645 (2014).
Chang, C. W. et al. Fecal microbiota transplantation prevents intestinal injury, upregulation of Toll-like receptors, and 5-fluorouracil/oxaliplatin-induced toxicity in colorectal cancer. Int. J. Mol. Sci. 21, 386 (2020).
Cui, M. et al. Faecal microbiota transplantation protects against radiation-induced toxicity. EMBO Mol. Med. 9, 448–461 (2017).
Xiao, H. W. et al. Gut microbiota-derived indole 3-propionic acid protects against radiation toxicity via retaining acyl-CoA-binding protein. Microbiome 8, 69 (2020).
Wang, F., Yin, Q., Chen, L. & Davis, M. M. Bifidobacterium can mitigate intestinal immunopathology in the context of CTLA-4 blockade. Proc. Natl Acad. Sci. USA 115, 157–161 (2018).
Budynek, P., Dabrowska, K., Skaradzinski, G. & Gorski, A. Bacteriophages and cancer. Arch. Microbiol. 192, 315–320 (2010).
Rasmussen, T. S. et al. Bacteriophage-mediated manipulation of the gut microbiome—promises and presents limitations. FEMS Microbiol. Rev. 44, 507–521 (2020).
Aykut, B. et al. The fungal mycobiome promotes pancreatic oncogenesis via activation of MBL. Nature 574, 264–267 (2019).
Durazzi, F. et al. Comparison between 16S rRNA and shotgun sequencing data for the taxonomic characterization of the gut microbiota. Sci. Rep. 11, 3030 (2021).
Huang, H. et al. Integrated analysis of microbiome and host transcriptome reveals correlations between gut microbiota and clinical outcomes in HBV-related hepatocellular carcinoma. Genome Med. 12, 102 (2020).
Moss, E. L., Maghini, D. G. & Bhatt, A. S. Complete, closed bacterial genomes from microbiomes using nanopore sequencing. Nat. Biotechnol. 38, 701–707 (2020).
Mallick, H. et al. Predictive metabolomic profiling of microbial communities using amplicon or metagenomic sequences. Nat. Commun. 10, 3136 (2019).
Zierer, J. et al. The fecal metabolome as a functional readout of the gut microbiome. Nat. Genet. 50, 790–795 (2018).
Garmaeva, S. et al. Studying the gut virome in the metagenomic era: challenges and perspectives. BMC Biol. 17, 84 (2019).
Vemuri, R., Shankar, E. M., Chieppa, M., Eri, R. & Kavanagh, K. Beyond just bacteria: functional biomes in the gut ecosystem including virome, mycobiome, archaeome and helminths. Microorganisms 8, 483 (2020).
O’Dwyer, D. N., Dickson, R. P. & Moore, B. B. The lung microbiome, immunity, and the pathogenesis of chronic lung disease. J. Immunol. 196, 4839–4847 (2016).
Dickson, R. P. & Huffnagle, G. B. The lung microbiome: new principles for respiratory bacteriology in health and disease. PLoS Pathog. 11, e1004923 (2015).
Schommer, N. N. & Gallo, R. L. Structure and function of the human skin microbiome. Trends Microbiol. 21, 660–668 (2013).
Ma, B., Forney, L. J. & Ravel, J. Vaginal microbiome: rethinking health and disease. Annu. Rev. Microbiol. 66, 371–389 (2012).
Pflughoeft, K. J. & Versalovic, J. Human microbiome in health and disease. Annu. Rev. Pathol. 7, 99–122 (2012).
Besser, J. et al. Next-generation sequencing technologies and their application to the study and control of bacterial infections. Clin. Microbiol. Infect. 24, 335–341 (2018).
Gosiewski, T. et al. Comprehensive detection and identification of bacterial DNA in the blood of patients with sepsis and healthy volunteers using next-generation sequencing method-the observation of DNAemia. Eur. J. Clin. Microbiol Infect. Dis. 36, 329–336 (2017).
Laurence, M., Hatzis, C. & Brash, D. E. J. P. O. Common contaminants in next-generation sequencing that hinder discovery of low-abundance microbes. PLoS ONE 9, e97876 (2014).
Eisenhofer, R. et al. Contamination in low microbial biomass microbiome studies: issues and recommendations. Trends Microbiol. 27, 105–117 (2019).
Glassing, A., Dowd, S. E., Galandiuk, S., Davis, B. & Chiodini, R. J. Inherent bacterial DNA contamination of extraction and sequencing reagents may affect interpretation of microbiota in low bacterial biomass samples. Gut Pathog. 8, 24 (2016).
Colgan, D. F. & Manley, J. L. Mechanism and regulation of mRNA polyadenylation. Genes Dev. 11, 2755–2766 (1997).
Sender, R., Fuchs, S. & Milo, R. Revised estimates for the number of human and bacteria cells in the body. PLoS Biol. 14, e1002533 (2016).
Davis, N. M., Proctor, D. M., Holmes, S. P., Relman, D. A. & Callahan, B. J. Simple statistical identification and removal of contaminant sequences in marker-gene and metagenomics data. Microbiome 6, 226 (2018).
Barrett, M., Hand, C. K., Shanahan, F., Murphy, T. & O’Toole, P. W. Mutagenesis by microbe: the role of the microbiota in shaping the cancer genome. Trends Cancer 6, 277–287 (2020).
He, Z. et al. Campylobacter jejuni promotes colorectal tumorigenesis through the action of cytolethal distending toxin. Gut 68, 289–300 (2019).
Dejea, C. M. et al. Patients with familial adenomatous polyposis harbor colonic biofilms containing tumorigenic bacteria. Science 359, 592–597 (2018).
Gonzalez-Cao, M. et al. Human endogenous retroviruses and cancer. Cancer Biol. Med. 13, 483–488 (2016).
Cherkasova, E. et al. Detection of an immunogenic HERV-E envelope with selective expression in clear cell kidney cancer. Cancer Res. 76, 2177–2185 (2016).
Kraus, B. et al. Vaccination directed against the human endogenous retrovirus-K envelope protein inhibits tumor growth in a murine model system. PLoS ONE 8, e72756 (2013).
Suh, Y., Amelio, I., Guerrero Urbano, T. & Tavassoli, M. Clinical update on cancer: molecular oncology of head and neck cancer. Cell Death Dis. 5, e1018 (2014).
Yin, H., Qu, J., Peng, Q. & Gan, R. Molecular mechanisms of EBV-driven cell cycle progression and oncogenesis. Med. Microbiol. Immunol. 208, 573–583 (2019).
Cheng, A. S. et al. Helicobacter pylori causes epigenetic dysregulation of FOXD3 to promote gastric carcinogenesis. Gastroenterology 144, 122–133 (2013).
Inamura, K. Colorectal cancers: an update on their molecular pathology. Cancers 10, 1390–1415 (2018).
Kopp, M., Durr, K., Steigleder, M., Clavel, T. & Rychlik, M. Development of stable isotope dilution assays for the quantitation of intra- and extracellular folate patterns of Bifidobacterium adolescentis. J. Chromatogr. A 1469, 48–59 (2016).
Lee, J. A. et al. Differential immune microenvironmental features of microsatellite-unstable colorectal cancers according to Fusobacterium nucleatum status. Cancer Immunol. Immunother. 70, 47–59 (2021).
Mima, K. et al. Fusobacterium nucleatum and T cells in colorectal carcinoma. JAMA Oncol. 1, 653–661 (2015).
Lopes, A. et al. Colibactin-positive Escherichia coli induce a procarcinogenic immune environment leading to immunotherapy resistance in colorectal cancer. Int. J. Cancer 146, 3147–3159 (2020).
Ling, Z. et al. Regulatory T cells and plasmacytoid dendritic cells within the tumor microenvironment in gastric cancer are correlated with gastric microbiota dysbiosis: a preliminary study. Front. Immunol. 10, 533 (2019).
Chen, T. et al. Fusobacterium nucleatum promotes M2 polarization of macrophages in the microenvironment of colorectal tumours via a TLR4-dependent mechanism. Cancer Immunol. Immunother. 67, 1635–1646 (2018).
Deng, Y. et al. TLR1/TLR2 signaling blocks the suppression of monocytic myeloid-derived suppressor cell by promoting its differentiation into M1-type macrophage. Mol. Immunol. 112, 266–273 (2019).
Kim, J. H., Kordahi, M. C., Chac, D. & DePaolo, R. W. Toll-like receptor-6 signaling prevents inflammation and impacts composition of the microbiota during inflammation-induced colorectal cancer. Cancer Prev. Res. 13, 25–40 (2020).
Hoste, E. et al. Innate sensing of microbial products promotes wound-induced skin cancer. Nat. Commun. 6, 5932 (2015).
Shi, Y. et al. Intratumoral accumulation of gut microbiota facilitates CD47-based immunotherapy via STING signaling. J. Exp. Med. 217, e20192282 (2020).
Maisonneuve, C. et al. Nod1 promotes colorectal carcinogenesis by regulating the immunosuppressive functions of tumor-infiltrating myeloid cells. Cell Rep. 34, 108677 (2021).
da Silva Correia, J. et al. Nod1-dependent control of tumor growth. Proc. Natl Acad. Sci. USA 103, 1840–1845 (2006).
Nguyen, T. T. et al. Lithocholic acid stimulates IL-8 expression in human colorectal cancer cells via activation of Erk1/2 MAPK and suppression of STAT3 activity. J. Cell. Biochem. 118, 2958–2967 (2017).
Baek, M. K. et al. Lithocholic acid upregulates uPAR and cell invasiveness via MAPK and AP-1 signaling in colon cancer cells. Cancer Lett. 290, 123–128 (2010).
Flynn, C. et al. Deoxycholic acid promotes the growth of colonic aberrant crypt foci. Mol. Carcinog. 46, 60–70 (2007).
Yao, L. et al. Acetate promotes SNAI1 expression by ACSS2-mediated histone acetylation under glucose limitation in renal cell carcinoma cell. Biosci. Rep. 40, BSR20200382 (2020).
Rojo, D. et al. Ranking the impact of human health disorders on gut metabolism: systemic lupus erythematosus and obesity as study cases. Sci. Rep. 5, 8310 (2015).
Ma, C. et al. Gut microbiome-mediated bile acid metabolism regulates liver cancer via NKT cells. Science 360, eaan5931 (2018).
Singh, N. et al. Activation of Gpr109a, receptor for niacin and the commensal metabolite butyrate, suppresses colonic inflammation and carcinogenesis. Immunity 40, 128–139 (2014).
Cuevas-Ramos, G. et al. Escherichia coli induces DNA damage in vivo and triggers genomic instability in mammalian cells. Proc. Natl Acad. Sci. USA 107, 11537–11542 (2010).
Jinadasa, R. N., Bloom, S. E., Weiss, R. S. & Duhamel, G. E. Cytolethal distending toxin: a conserved bacterial genotoxin that blocks cell cycle progression, leading to apoptosis of a broad range of mammalian cell lineages. Microbiol. 157, 1851–1875 (2011).
Maddocks, O. D., Scanlon, K. M. & Donnenberg, M. S. An Escherichia coli effector protein promotes host mutation via depletion of DNA mismatch repair proteins. mBio 4, e00152–00113 (2013).
Santos, J. C. et al. Helicobacter pylori infection modulates the expression of miRNAs associated with DNA mismatch repair pathway. Mol. Carcinog. 56, 1372–1379 (2017).
Kim, J. J. et al. Helicobacter pylori impairs DNA mismatch repair in gastric epithelial cells. Gastroenterology 123, 542–553 (2002).
Chung, L. et al. Bacteroides fragilis toxin coordinates a pro-carcinogenic inflammatory cascade via targeting of colonic epithelial cells. Cell Host Microbe 23, 203–214 (2018).
Wang, F., Meng, W., Wang, B. & Qiao, L. Helicobacter pylori-induced gastric inflammation and gastric cancer. Cancer Lett. 345, 196–202 (2014).
Rubinstein, M. R. et al. Fusobacterium nucleatum promotes colorectal carcinogenesis by modulating E-cadherin/beta-catenin signaling via its FadA adhesin. Cell Host Microbe 14, 195–206 (2013).
Rossi, T. et al. Microbiota-derived metabolites in tumor progression and metastasis. Int. J. Mol. Sci. 21, 5786 (2020).
Yoshimoto, S. et al. Obesity-induced gut microbial metabolite promotes liver cancer through senescence secretome. Nature 499, 97–101 (2013).
Seifert, L. et al. The necrosome promotes pancreatic oncogenesis via CXCL1 and Mincle-induced immune suppression. Nature 532, 245–249 (2016).
Vitiello, G. A., Cohen, D. J. & Miller, G. Harnessing the microbiome for pancreatic cancer immunotherapy. Trends Cancer 5, 670–676 (2019).
Das, S., Shapiro, B., Vucic, E. A., Vogt, S. & Bar-Sagi, D. Tumor cell-derived IL1β promotes desmoplasia and immune suppression in pancreatic cancer. Cancer Res. 80, 1088–1101 (2020).
Pushalkar, S. et al. The pancreatic cancer microbiome promotes oncogenesis by induction of innate and adaptive immune suppression. Cancer Discov. 8, 403–416 (2018).
Cao, S. et al. Dynamic host immune response in virus-associated cancers. Commun. Biol. 2, 109 (2019).
Overacre-Delgoffe, A. E. et al. Microbiota-specific T follicular helper cells drive tertiary lymphoid structures and anti-tumor immunity against colorectal cancer. Immunity 54, 2812–2824 (2021).
Wieland, A. et al. Defining HPV-specific B cell responses in patients with head and neck cancer. Nature 597, 274–278 (2021).
Cillo, A. R. et al. Immune landscape of viral- and carcinogen-driven head and neck cancer. Immunity 52, 183–199 (2020).
Kalaora, S. et al. Identification of bacteria-derived HLA-bound peptides in melanoma. Nature 592, 138–143 (2021).
McQuade, J. L., Daniel, C. R., Helmink, B. A. & Wargo, J. A. Modulating the microbiome to improve therapeutic response in cancer. Lancet Oncol. 20, e77–e91 (2019).
Maleki, S. et al. P864 Combination of fecal microbiota transplantation from healthy donors with anti-PD1 immunotherapy in treatment-naïve advanced or metastatic melanoma patients. J. Immunother. Cancer 8, A11–A12 (2020).
Zhao, Y. et al. Safety and efficacy of fecal microbiota transplantation for grade IV steroid refractory GI-GvHD patients: interim results from FMT2017002 trial. Front. Immunol. 12, 678476 (2021).
Giles, E. M., D’Adamo, G. L. & Forster, S. C. The future of faecal transplants. Nat. Rev. Microbiol. 17, 719 (2019).
Dizman, N. et al. Nivolumab plus ipilimumab with or without live bacterial supplementation in metastatic renal cell carcinoma: a randomized phase 1 trial. Nat. Med. https://doi.org/10.1038/s41591-022-01694-6 (2022).
Laute-Caly, D. L. et al. The flagellin of candidate live biotherapeutic Enterococcus gallinarum MRx0518 is a potent immunostimulant. Sci. Rep. 9, 801 (2019).
Tanoue, T. et al. A defined commensal consortium elicits CD8 T cells and anti-cancer immunity. Nature 565, 600–605 (2019).
Feuerstadt, P. et al. SER-109, an oral microbiome therapy for recurrent Clostridioides difficile infection. N. Engl. J. Med. 386, 220–229 (2022).
Lemon, K. P., Armitage, G. C., Relman, D. A. & Fischbach, M. A. Microbiota-targeted therapies: an ecological perspective. Sci. Transl. Med. 4, 137rv135 (2012).
Helmink, B. A. et al. B cells and tertiary lymphoid structures promote immunotherapy response. Nature 577, 549–555 (2020).
Cabrita, R. et al. Tertiary lymphoid structures improve immunotherapy and survival in melanoma. Nature 577, 561–565 (2020).
Petitprez, F. et al. B cells are associated with survival and immunotherapy response in sarcoma. Nature 577, 556–560 (2020).
Pflug, N. et al. Efficacy of antineoplastic treatment is associated with the use of antibiotics that modulate intestinal microbiota. Oncoimmunology 5, e1150399 (2016).
Weber, D. et al. Microbiota disruption induced by early use of broad-spectrum antibiotics is an independent risk factor of outcome after allogeneic stem cell transplantation. Biol. Blood Marrow Transplant. 23, 845–852 (2017).
Nenclares, P. et al. Impact of antibiotic use during curative treatment of locally advanced head and neck cancers with chemotherapy and radiotherapy. Eur. J. Cancer 131, 9–15 (2020).
Selle, K. et al. In vivo targeting of Clostridioides difficile using phage-delivered CRISPR–Cas3 antimicrobials. mBio 11, e00019–e00020 (2020).
Chen, P. B. et al. Directed remodeling of the mouse gut microbiome inhibits the development of atherosclerosis. Nat. Biotechnol. 38, 1288–1297 (2020).
Kroemer, G., Lopez-Otin, C., Madeo, F. & de Cabo, R. Carbotoxicity-noxious effects of carbohydrates. Cell 175, 605–614 (2018).
Buque, A. et al. Immunoprophylactic and immunotherapeutic control of hormone receptor-positive breast cancer. Nat. Commun. 11, 3819 (2020).
Ferrere, G. et al. Ketogenic diet and ketone bodies enhance the anticancer effects of PD-1 blockade. JCI Insight 6, e145207 (2021).
Levesque, S. et al. A synergistic triad of chemotherapy, immune checkpoint inhibitors, and caloric restriction mimetics eradicates tumors in mice. Oncoimmunology 8, e1657375 (2019).
Pietrocola, F. et al. Caloric restriction mimetics enhance anticancer immunosurveillance. Cancer Cell 30, 147–160 (2016).
Pomatto-Watson, L. C. D. et al. Daily caloric restriction limits tumor growth more effectively than caloric cycling regardless of dietary composition. Nat. Commun. 12, 6201 (2021).
Wang, Y. et al. NAD+ supplement potentiates tumor-killing function by rescuing defective TUB-mediated NAMPT transcription in tumor-infiltrated T cells. Cell Rep. 36, 109516 (2021).
Baruch, E. N., Wang, J. & Wargo, J. A. Gut microbiota and antitumor immunity: potential mechanisms for clinical effect. Cancer Immunol. Res. 9, 365–370 (2021).
Becerril-Alarcon, Y. et al. Inulin supplementation reduces systolic blood pressure in women with breast cancer undergoing neoadjuvant chemotherapy. Cardiovasc. Ther. 2019, 5707150 (2019).
Fogliano, V. et al. In vitro bioaccessibility and gut biotransformation of polyphenols present in the water-insoluble cocoa fraction. Mol. Nutr. Food Res. 55, S44–S55 (2011).
Scott, K. P., Martin, J. C., Duncan, S. H. & Flint, H. J. Prebiotic stimulation of human colonic butyrate-producing bacteria and bifidobacteria, in vitro. FEMS Microbiol. Ecol. 87, 30–40 (2014).
Sanders, M. E., Merenstein, D. J., Reid, G., Gibson, G. R. & Rastall, R. A. Probiotics and prebiotics in intestinal health and disease: from biology to the clinic. Nat. Rev. Gastroenterol. Hepatol. 16, 605–616 (2019).
Jessy, T. Immunity over inability: the spontaneous regression of cancer. J. Nat. Sci. Biol. Med. 2, 43–49 (2011).
Kucerova, P. & Cervinkova, M. Spontaneous regression of tumour and the role of microbial infection–possibilities for cancer treatment. Anticancer Drugs 27, 269–277 (2016).
Coley, W. B. The treatment of inoperable sarcoma with the’mixed toxins of erysipelas and bacillus prodigiosus.: immediate and final results in one hundred and forty cases. Proc. R. Soc. Med. 31, 456–465 (1898).
Rook, G. Tumours and Coley’s toxins. Nature 357, 545 (1992).
Andtbacka, R. H. et al. Talimogene laherparepvec improves durable response rate in patients with advanced melanoma. J. Clin. Oncol. 33, 2780–2788 (2015).
Roberts, N. J. et al. Intratumoral injection of Clostridium novyi-NT spores induces antitumor responses. Sci. Transl. Med. 6, 249ra111 (2014).
Kelly, C. M. et al. Objective response rate among patients with locally advanced or metastatic sarcoma treated with talimogene laherparepvec in combination with pembrolizumab: a phase 2 clinical trial. JAMA Oncol. 6, 402–408 (2020).
Rha, S. Y. et al. Abstract CT121: a phase Ib study of recombinant vaccinia virus in combination with immune checkpoint inhibition (ICI) in advanced renal cell carcinoma (RCC). Cancer Res. 80, CT121–CT121 (2020).
Krupar, R. et al. Abstract LB-095: HPV E7 antigen-expressing listeria-based immunotherapy (ADXS11-001) prior to robotic surgery for HPV-positive oropharyngeal cancer enhances HPV-specific T cell immunity. Cancer Res. 76, LB-095–LB-095 (2016).
Safran, H. et al. ADXS11-001 Lm-LLO immunotherapy, mitomycin, 5-fluorouracil (5-FU) and intensity-modulated radiation therapy (IMRT) for anal cancer. J. Clin. Oncol. 35, e15072–e15072 (2017).
van Zyl, D. G., Mautner, J. & Delecluse, H. J. Progress in EBV vaccines. Front Oncol. 9, 104 (2019).
Sedighi, M. et al. Therapeutic bacteria to combat cancer; current advances, challenges, and opportunities. Cancer Med. 8, 3167–3181 (2019).
Giannakis, M. et al. Genomic correlates of immune-cell infiltrates in colorectal carcinoma. Cell Rep. 15, 857–865 (2016).
Chen, P.-L. et al. Analysis of immune signatures in longitudinal tumor samples yields insight into biomarkers of response and mechanisms of resistance to immune checkpoint blockade. Cancer Discov. 6, 827–837 (2016).
Tumeh, P. C. et al. PD-1 blockade induces responses by inhibiting adaptive immune resistance. Nature 515, 568–571 (2014).
Topalian, S. L. et al. Safety, activity, and immune correlates of anti-PD-1 antibody in cancer. N. Engl. J. Med. 366, 2443–2454 (2012).
Bronkhorst, A. J., Ungerer, V. & Holdenrieder, S. The emerging role of cell-free DNA as a molecular marker for cancer management. Biomol. Detect. Quantif. 17, 100087 (2019).
Halley, A. et al. The role of the microbiome in cancer and therapy efficacy: focus on lung cancer. Anticancer Res. 40, 4807–4818 (2020).
Oliva, M. et al. Immune biomarkers of response to immune-checkpoint inhibitors in head and neck squamous cell carcinoma. Ann. Oncol. 30, 57–67 (2019).
Temraz, S. et al. Gut microbiome: a promising biomarker for immunotherapy in colorectal cancer. Int. J. Mol. Sci. 20, 4155 (2019).
van Poelgeest, M. I. et al. Potential use of lymph node-derived HPV-specific T cells for adoptive cell therapy of cervical cancer. Cancer Immunol. Immunother. 65, 1451–1463 (2016).
Menzel, A. et al. Common and novel markers for measuring inflammation and oxidative stress ex vivo in research and clinical practice—which to use regarding disease outcomes? Antioxidants 10, 414 (2021).
Howard, R., Kanetsky, P. A. & Egan, K. M. Exploring the prognostic value of the neutrophil-to-lymphocyte ratio in cancer. Sci. Rep. 9, 19673 (2019).
Cordoba, R., Eyre, T. A., Klepin, H. D., Wildes, T. M. & Goede, V. A comprehensive approach to therapy of haematological malignancies in older patients. Lancet Haematol. 8, e840–e852 (2021).
Zhang, H. et al. Novel immune infiltrating cell signature based on cell pair algorithm is a prognostic marker in cancer. Front. Immunol. 12, 694490 (2021).
Nixon, A. B. et al. Peripheral immune-based biomarkers in cancer immunotherapy: can we realize their predictive potential? J. Immunother. Cancer 7, 325 (2019).
Verma, M., Hontecillas, R., Tubau-Juni, N., Abedi, V. & Bassaganya-Riera, J. Challenges in personalized nutrition and health. Front Nutr. 5, 117 (2018).
Kalantar-Zadeh, K. et al. A human pilot trial of ingestible electronic capsules capable of sensing different gases in the gut. Nat. Electron. 1, 79–87 (2018).
Jalili-Firoozinezhad, S. et al. A complex human gut microbiome cultured in an anaerobic intestine-on-a-chip. Nat. Biomed. Eng. 3, 520–531 (2019).
Cammarota, G. et al. Gut microbiome, big data and machine learning to promote precision medicine for cancer. Nat. Rev. Gastroenterol. Hepatol. 17, 635–648 (2020).
Zeng, T., Yu, X. & Chen, Z. Applying artificial intelligence in the microbiome for gastrointestinal diseases: a review. J. Gastroenterol. Hepatol. 36, 832–840 (2021).
Stylianopoulos, T., Munn, L. L. & Jain, R. K. Reengineering the physical microenvironment of tumors to improve drug delivery and efficacy: from mathematical modeling to bench to bedside. Trends Cancer 4, 292–319 (2018).
Bucci, V. & Xavier, J. B. Towards predictive models of the human gut microbiome. J. Mol. Biol. 426, 3907–3916 (2014).
Karlsson, F. H., Nookaew, I., Petranovic, D. & Nielsen, J. Prospects for systems biology and modeling of the gut microbiome. Trends Biotechnol. 29, 251–258 (2011).
Dourson, M. et al. The future of uncertainty factors with in vitro studies using human cells. Toxicol. Sci. 186, 12–17 (2022).
Acknowledgements
The authors thank R. G. Witt, S. Cass, R. Traweek and E. Baruch for their thoughtful input and assistance with preparation of this manuscript. L.Z. and G.K. are supported by Seerave Foundation, the RHU Immunolife, ANR Ileobiome (19-CE15-0029-01), the Ligue contre le Cancer (équipe labellisée); Agence National de la Recherche (ANR)—Projets blancs; AMMICa (US23/CNRS UMS3655); Association pour la recherche sur le cancer (ARC); Cancéropôle Ile-de-France; Fondation pour la Recherche Médicale (FRM); a donation by Elior; Equipex Onco-Pheno-Screen; European Joint Programme on Rare Diseases (EJPRD); Gustave Roussy Odyssea, the European Union Horizon 2020 Projects Oncobiome and Crimson; Fondation Carrefour; Institut National du Cancer (INCa); Institut Universitaire de France; LabEx Immuno-Oncology (ANR-18-IDEX-0001); a Cancer Research ASPIRE Award from the Mark Foundation; SIRIC Stratified Oncology Cell DNA Repair and Tumor Immune Elimination (SOCRATE); and SIRIC Cancer Research and Personalized Medicine (CARPEM). This study contributes to the IdEx Université de Paris, ANR-18-IDEX-0001. E.M.P. was supported by the CPRIT Training Award (RP210028).
Author information
Authors and Affiliations
Contributions
All authors contributed equally to the design, preparation and review of this manuscript. J.A.W. supervised and oversaw all aspects of this work.
Corresponding author
Ethics declarations
Competing interests
J.A.W. is an inventor on a US patent application (PCT/US17/53.717); reports compensation for speaker’s bureau and honoraria from Imedex, Dava Oncology, Omniprex, Illumina, Gilead, PeerView, MedImmune and Bristol-Myers Squibb; serves as a consultant/advisory board member for Roche/Genentech, Novartis, AstraZeneca, GlaxoSmithKline, Bristol-Myers Squibb, Merck, Biothera Pharmaceuticals and Micronoma. J.A.W. holds stock options from Micronoma. L.Z. received research contracts from Kaleido, Innovate Pharma and Pilege. G.K. had a research contract with Kaleido. L.Z is and G.K. was among the scientific cofounders of EverImmune, a biotech company devoted to the use of commensal bacteria for the treatment of cancers.
Peer review
Peer review information
Nature Medicine thanks the anonymous reviewers for their contribution to the peer review of this work. Primary Handling Editor: Karen O’Leary, in collaboration with the Nature Medicine team.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Data 1
Supplementary Table 1
Rights and permissions
About this article
Cite this article
Park, E.M., Chelvanambi, M., Bhutiani, N. et al. Targeting the gut and tumor microbiota in cancer. Nat Med 28, 690–703 (2022). https://doi.org/10.1038/s41591-022-01779-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41591-022-01779-2
This article is cited by
-
Effects of dietary intervention on human diseases: molecular mechanisms and therapeutic potential
Signal Transduction and Targeted Therapy (2024)
-
Cardiovascular disease and cancer: shared risk factors and mechanisms
Nature Reviews Cardiology (2024)
-
The effect of physical exercise on anticancer immunity
Nature Reviews Immunology (2024)
-
Personalizing Oncolytic Immunovirotherapy Approaches
Molecular Diagnosis & Therapy (2024)
-
Modulation of intestinal metabolites by calorie restriction and its association with gut microbiota in a xenograft model of colorectal cancer
Discover Oncology (2024)