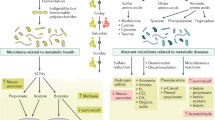
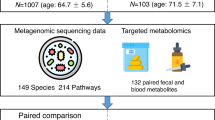
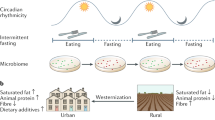
Abstract
Cardiometabolic diseases have become a leading cause of morbidity and mortality globally. They have been tightly linked to microbiome taxonomic and functional composition, with diet possibly mediating some of the associations described. Both the microbiome and diet are modifiable, which opens the way for novel therapeutic strategies. High-throughput omics techniques applied on microbiome samples (meta-omics) hold the unprecedented potential to shed light on the intricate links between diet, the microbiome, the metabolome and cardiometabolic health, with a top-down approach. However, effective integration of complementary meta-omic techniques is an open challenge and their application on large cohorts is still limited. Here we review meta-omics techniques and discuss their potential in this context, highlighting recent large-scale efforts and the novel insights they provided. Finally, we look to the next decade of meta-omics research and discuss various translational and clinical pathways to improving cardiometabolic health.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
References
Mathers, C. D. & Loncar, D. Projections of global mortality and burden of disease from 2002 to 2030. PLoS Med. 3, e442 (2006).
National Academies of Sciences, Engineering, and Medicine et al. in High and Rising Mortality Rates Among Working-Age Adults Ch. 9 (National Academies Press, 2021).
Jagannathan, R., Patel, S. A., Ali, M. K. & Narayan, K. M. V. Global updates on cardiovascular disease mortality trends and attribution of traditional risk factors. Curr. Diab. Rep. 19, 44 (2019).
Korecka, A. & Arulampalam, V. The gut microbiome: scourge, sentinel or spectator? J. Oral Microbiol. 4, https://doi.org/10.3402/jom.v4i0.9367 (2012).
Tang, W. H. W. & Hazen, S. L. The gut microbiome and its role in cardiovascular diseases. Circulation 135, 1008–1010 (2017).
Menni, C. et al. Gut microbial diversity is associated with lower arterial stiffness in women. Eur. Heart J. 39, 2390–2397 (2018).
Nogal, A., Valdes, A. M. & Menni, C. The role of short-chain fatty acids in the interplay between gut microbiota and diet in cardio-metabolic health. Gut Microbes 13, 1–24 (2021).
Hansen, T. H., Gøbel, R. J., Hansen, T. & Pedersen, O. The gut microbiome in cardio-metabolic health. Genome Med. 7, 33 (2015).
Jardon, K. M., Canfora, E. E., Goossens, G. H. & Blaak, E. E. Dietary macronutrients and the gut microbiome: a precision nutrition approach to improve cardiometabolic health. Gut 71, 1214–1226 (2022).
Wan, Y. et al. Contribution of diet to gut microbiota and related host cardiometabolic health: diet–gut interaction in human health. Gut Microbes 11, 603–609 (2020).
Karlsson, F. H. et al. Gut metagenome in European women with normal, impaired and diabetic glucose control. Nature 498, 99–103 (2013).
Talmor-Barkan, Y. et al. Metabolomic and microbiome profiling reveals personalized risk factors for coronary artery disease. Nat. Med. 28, 295–302 (2022).
Sumida, K. et al. Circulating microbiota in cardiometabolic disease. Front. Cell. Infect. Microbiol. 12, 892232 (2022).
Brunius, C., Shi, L. & Landberg, R. Metabolomics for improved understanding and prediction of cardiometabolic diseases—recent findings from human studies. Curr. Nutr. Rep. 4, 348–364 (2015).
Johnson, M. Diet and nutrition: implications to cardiometabolic health. J. Cardiol. Cardiovasc. Sci. 3, 4–9 (2019).
Doran, S. et al. Multi-omics approaches for revealing the complexity of cardiovascular disease. Brief. Bioinformatics 22, bbab061 (2021).
Joshi, A., Rienks, M., Theofilatos, K. & Mayr, M. Systems biology in cardiovascular disease: a multiomics approach. Nat. Rev. Cardiol. 18, 313–330 (2020).
Abu-Ali, G. S. et al. Metatranscriptome of human faecal microbial communities in a cohort of adult men. Nat. Microbiol. 3, 356–366 (2018).
Schirmer, M. et al. Dynamics of metatranscription in the inflammatory bowel disease gut microbiome. Nat. Microbiol. 3, 337–346 (2018).
Zierer, J. et al. The fecal metabolome as a functional readout of the gut microbiome. Nat. Genet. 50, 790–795 (2018).
Quince, C., Walker, A. W., Simpson, J. T., Loman, N. J. & Segata, N. Shotgun metagenomics, from sampling to analysis. Nat. Biotechnol. 35, 833–844 (2017).
Martinez, K. B., Leone, V. & Chang, E. B. Microbial metabolites in health and disease: navigating the unknown in search of function. J. Biol. Chem. 292, 8553–8559 (2017).
Douglas, G. M. et al. PICRUSt2 for prediction of metagenome functions. Nat. Biotechnol. 38, 685–688 (2020).
Shakya, M., Lo, C.-C. & Chain, P. S. G. Advances and challenges in metatranscriptomic analysis. Front. Genet. 10, 904 (2019).
Valles-Colomer, M. et al. Meta-omics in inflammatory bowel disease research: applications, challenges, and guidelines. J. Chrons Colitis 10, 735–746 (2016).
Kleiner, M. Metaproteomics: much more than measuring gene expression in microbial communities. mSystems 4, e00115-19 (2019).
Roberts, L. D., Souza, A. L., Gerszten, R. E. & Clish, C. B. Targeted metabolomics. Curr. Protoc. Mol. Biol. 98, 30.2.1–30.2.24 (2012).
Menni, C., Zierer, J., Valdes, A. M. & Spector, T. D. Mixing omics: combining genetics and metabolomics to study rheumatic diseases. Nat. Rev. Rheumatol. 13, 174–181 (2017).
Kuleš, J. et al. Combined untargeted and targeted metabolomics approaches reveal urinary changes of amino acids and energy metabolism in canine babesiosis with different levels of kidney function. Front. Microbiol. 12, 715701 (2021).
Hollywood, K., Brison, D. R. & Goodacre, R. Metabolomics: current technologies and future trends. Proteomics 6, 4716–4723 (2006).
Linnarsson, S. & Teichmann, S. A. Single-cell genomics: coming of age. Genome Biol. 17, 97 (2016).
Pasolli, E. et al. Extensive unexplored human microbiome diversity revealed by over 150,000 genomes from metagenomes spanning age, geography, and lifestyle. Cell 176, 649–662.e20 (2019).
Almeida, A. et al. A unified catalog of 204,938 reference genomes from the human gut microbiome. Nat. Biotechnol. 39, 105–114 (2021).
Lloréns-Rico, V., Simcock, J. A., Huys, G. R. B. & Raes, J. Single-cell approaches in human microbiome research. Cell 185, 2725–2738 (2022).
Lagier, J.-C. et al. Culturing the human microbiota and culturomics. Nat. Rev. Microbiol. 16, 540–550 (2018).
Van de Wiele, T., Van den Abbeele, P., Ossieur, W., Possemiers, S. & Marzorati, M. in The Impact of Food Bioactives on Health: In Vitro and Ex Vivo Models 305–317 (Springer International Publishing, 2015).
Minot, S. et al. The human gut virome: inter-individual variation and dynamic response to diet. Genome Res. 21, 1616–1625 (2011).
Garmaeva, S. et al. Stability of the human gut virome and effect of gluten-free diet. Cell Rep. 35, 109132 (2021).
Scarpellini, E. et al. The human gut microbiota and virome: potential therapeutic implications. Dig. Liver Dis. 47, 1007–1012 (2015).
Warmbrunn, M. V. et al. Gut microbiota: a promising target against cardiometabolic diseases. Expert Rev. Endocrinol. Metab. 15, 13–27 (2020).
Herold, M. et al. Integration of time-series meta-omics data reveals how microbial ecosystems respond to disturbance. Nat. Commun. 11, 5281 (2020).
Falony, G. et al. Population-level analysis of gut microbiome variation. Science 352, 560–564 (2016).
Zhernakova, A. et al. Population-based metagenomics analysis reveals markers for gut microbiome composition and diversity. Science 352, 565–569 (2016).
Fromentin, S. et al. Microbiome and metabolome features of the cardiometabolic disease spectrum. Nat. Med. 28, 303–314 (2022).
Asnicar, F. et al. Microbiome connections with host metabolism and habitual diet from 1,098 deeply phenotyped individuals. Nat. Med. 27, 321–332 (2021).
Wilmes, P., Heintz-Buschart, A. & Bond, P. L. A decade of metaproteomics: where we stand and what the future holds. Proteomics 15, 3409–3417 (2015).
Lloyd-Price, J. et al. Multi-omics of the gut microbial ecosystem in inflammatory bowel diseases. Nature 569, 655–662 (2019).
Zhou, W. et al. Longitudinal multi-omics of host–microbe dynamics in prediabetes. Nature 569, 663–671 (2019).
Zhang, Y. et al. Discovery of bioactive microbial gene products in inflammatory bowel disease. Nature 606, 754–760 (2022).
Oliveira, P. H. Bacterial epigenomics: coming of age. mSystems 6, e0074721 (2021).
Hiraoka, S. et al. Metaepigenomic analysis reveals the unexplored diversity of DNA methylation in an environmental prokaryotic community. Nat. Commun. 10, 159 (2019).
Singh, R. K. et al. Influence of diet on the gut microbiome and implications for human health. J. Transl. Med. 15, 73 (2017).
Rothschild, D. et al. Environment dominates over host genetics in shaping human gut microbiota. Nature 555, 210–215 (2018).
Tramontano, M. et al. Nutritional preferences of human gut bacteria reveal their metabolic idiosyncrasies. Nat. Microbiol. 3, 514–522 (2018).
Cummings, J. H. & Macfarlane, G. T. The control and consequences of bacterial fermentation in the human colon. J. Appl. Bacteriol. 70, 443–459 (1991).
Vieira-Silva, S. et al. Species–function relationships shape ecological properties of the human gut microbiome. Nat. Microbiol. 1, 16088 (2016).
Fehlner-Peach, H. et al. Distinct polysaccharide utilization profiles of human intestinal Prevotella copri isolates. Cell Host Microbe 26, 680–690.e5 (2019).
Wu, G. D. et al. Linking long-term dietary patterns with gut microbial enterotypes. Science 334, 105–108 (2011).
Walker, A. W. et al. Dominant and diet-responsive groups of bacteria within the human colonic microbiota. ISME J. 5, 220–230 (2011).
Ley, R. E., Turnbaugh, P. J., Klein, S. & Gordon, J. I. Microbial ecology: human gut microbes associated with obesity. Nature 444, 1022–1023 (2006).
David, L. A. et al. Diet rapidly and reproducibly alters the human gut microbiome. Nature 505, 559–563 (2014).
Johnson, A. J. et al. Daily sampling reveals personalized diet–microbiome associations in humans. Cell Host Microbe 25, 789–802.e5 (2019).
Wang, D. D. et al. The gut microbiome modulates the protective association between a Mediterranean diet and cardiometabolic disease risk. Nat. Med. 27, 333–343 (2021).
Ferro-Luzzi, A. et al. Changing the Mediterranean diet: effects on blood lipids. Am. J. Clin. Nutr. 40, 1027–1037 (1984).
Ghosh, T. S. et al. Mediterranean diet intervention alters the gut microbiome in older people reducing frailty and improving health status: the NU-AGE 1-year dietary intervention across five European countries. Gut 69, 1218–1228 (2020).
Turpin, W. et al. Mediterranean-like dietary pattern associations with gut microbiome composition and subclinical gastrointestinal inflammation. Gastroenterology 163, 685–698 (2022).
Nakayama, J. et al. Impact of Westernized diet on gut microbiota in children on Leyte Island. Front. Microbiol. 8, 197 (2017).
Tett, A. et al. The Prevotella copri complex comprises four distinct clades underrepresented in Westernized populations. Cell Host Microbe 26, 666–679.e7 (2019).
Kovatcheva-Datchary, P. et al. Dietary fiber-induced improvement in glucose metabolism is associated with increased abundance of Prevotella. Cell Metab. 22, 971–982 (2015).
Tett, A., Pasolli, E., Masetti, G., Ercolini, D. & Segata, N. Prevotella diversity, niches and interactions with the human host. Nat. Rev. Microbiol. 19, 585–599 (2021).
Meslier, V. et al. Mediterranean diet intervention in overweight and obese subjects lowers plasma cholesterol and causes changes in the gut microbiome and metabolome independently of energy intake. Gut 69, 1258–1268 (2020).
Ang, Q. Y. et al. Ketogenic diets alter the gut microbiome resulting in decreased intestinal TH17 cells. Cell 181, 1263–1275.e16 (2020).
Rondanelli, M. et al. The potential roles of very low calorie, very low calorie ketogenic diets and very low carbohydrate diets on the gut microbiota composition. Front. Endocrinol. 12, 662591 (2021).
Guo, Y. et al. Intermittent fasting improves cardiometabolic risk factors and alters gut microbiota in metabolic syndrome patients. J. Clin. Endocrinol. Metab. 106, 64–79 (2021).
Ratiner, K., Shapiro, H., Goldenberg, K. & Elinav, E. Time-limited diets and the gut microbiota in cardiometabolic disease. J. Diabetes 14, 377–393 (2022).
Attaye, I., van Oppenraaij, S., Warmbrunn, M. V. & Nieuwdorp, M. The role of the gut microbiota on the beneficial effects of ketogenic diets. Nutrients 14, 191 (2022).
Barabási, A.-L., Menichetti, G. & Loscalzo, J. The unmapped chemical complexity of our diet. Nat. Food 1, 33–37 (2019).
Clarke, R. J. Coffee: Chemistry Vol. 1 (Springer Science & Business Media, 2012).
Ruskovska, T., Maksimova, V. & Milenkovic, D. Polyphenols in human nutrition: from the in vitro antioxidant capacity to the beneficial effects on cardiometabolic health and related inter-individual variability—an overview and perspective. Br. J. Nutr. 123, 241–254 (2020).
Corrêa, T. A. F., Rogero, M. M., Hassimotto, N. M. A. & Lajolo, F. M. The two-way polyphenols–microbiota interactions and their effects on obesity and related metabolic diseases. Front. Nutr. 6, 188 (2019).
Cardona, F., Andrés-Lacueva, C., Tulipani, S., Tinahones, F. J. & Queipo-Ortuño, M. I. Benefits of polyphenols on gut microbiota and implications in human health. J. Nutr. Biochem. 24, 1415–1422 (2013).
Mompeo, O. et al. Consumption of stilbenes and flavonoids is linked to reduced risk of obesity independently of fiber intake. Nutrients 12, 1871 (2020).
Namazi, N., Irandoost, P., Larijani, B. & Azadbakht, L. The effects of supplementation with conjugated linoleic acid on anthropometric indices and body composition in overweight and obese subjects: a systematic review and meta-analysis. Crit. Rev. Food Sci. Nutr. 59, 2720–2733 (2019).
Chen, Y. et al. Orally administered CLA ameliorates DSS-induced colitis in mice via intestinal barrier improvement, oxidative stress reduction, and inflammatory cytokine and gut microbiota modulation. J. Agric. Food Chem. 67, 13282–13298 (2019).
Rosberg-Cody, E. et al. Recombinant lactobacilli expressing linoleic acid isomerase can modulate the fatty acid composition of host adipose tissue in mice. Microbiology 157, 609–615 (2011).
He, Y. et al. Metabolomic changes upon conjugated linoleic acid supplementation and predictions of body composition responsiveness. J. Clin. Endocrinol. Metab. 107, 2606–2615 (2022).
Valdes, A. M., Walter, J., Segal, E. & Spector, T. D. Role of the gut microbiota in nutrition and health. Brit. Med. J. 361, k2179 (2018).
Cryan, J. F. et al. The microbiota–gut–brain axis. Physiol. Rev. 99, 1877–2013 (2019).
Yoo, W. et al. High-fat diet-induced colonocyte dysfunction escalates microbiota-derived trimethylamine N-oxide. Science 373, 813–818 (2021).
Dodd, D. et al. A gut bacterial pathway metabolizes aromatic amino acids into nine circulating metabolites. Nature 551, 648–652 (2017).
Dekkers, K. F. et al. An online atlas of human plasma metabolite signatures of gut microbiome composition. Nat. Commun. 13, 5370 (2022).
Rath, S., Heidrich, B., Pieper, D. H. & Vital, M. Uncovering the trimethylamine-producing bacteria of the human gut microbiota. Microbiome 5, 54 (2017).
Thomas, A. M. et al. Metagenomic analysis of colorectal cancer datasets identifies cross-cohort microbial diagnostic signatures and a link with choline degradation. Nat. Med. 25, 667–678 (2019).
Falony, G., Vieira-Silva, S. & Raes, J. Microbiology meets big data: the case of gut microbiota-derived trimethylamine. Annu. Rev. Microbiol. 69, 305–321 (2015).
Cai, Y.-Y. et al. Integrated metagenomics identifies a crucial role for trimethylamine-producing Lachnoclostridium in promoting atherosclerosis. npj Biofilms Microbiomes 8, 11 (2022).
Schugar, R. C. et al. Gut microbe-targeted choline trimethylamine lyase inhibition improves obesity via rewiring of host circadian rhythms. eLife 11, e63998 (2022).
Louis, P. & Flint, H. J. Formation of propionate and butyrate by the human colonic microbiota. Environ. Microbiol. 19, 29–41 (2017).
Gasaly, N., Hermoso, M. A. & Gotteland, M. Butyrate and the fine-tuning of colonic homeostasis: implication for inflammatory bowel diseases. Int. J. Mol. Sci. 22, 3061 (2021).
Morrison, D. J. & Preston, T. Formation of short chain fatty acids by the gut microbiota and their impact on human metabolism. Gut Microbes 7, 189–200 (2016).
Valles-Colomer, M. et al. The neuroactive potential of the human gut microbiota in quality of life and depression. Nat. Microbiol. 4, 623–632 (2019).
Lai, Y. et al. High-coverage metabolomics uncovers microbiota-driven biochemical landscape of interorgan transport and gut–brain communication in mice. Nat. Commun. 12, –166000 (2021).
Lefort, C. & Cani, P. D. The liver under the spotlight: bile acids and oxysterols as pivotal actors controlling metabolism. Cells 10, 400 (2021).
Xie, A.-J., Mai, C.-T., Zhu, Y.-Z., Liu, X.-C. & Xie, Y. Bile acids as regulatory molecules and potential targets in metabolic diseases. Life Sci. 287, 120152 (2021).
De Vos, W. M., Tilg, H., Van Hul, M. & Cani, P. D. Gut microbiome and health: mechanistic insights. Gut 71, 1020–1032 (2022).
De Aguiar Vallim, T. Q., Tarling, E. J. & Edwards, P. A. Pleiotropic roles of bile acids in metabolism. Cell Metab. 17, 657–669 (2013).
Sato, Y. et al. Novel bile acid biosynthetic pathways are enriched in the microbiome of centenarians. Nature 599, 458–464 (2021).
Tomasova, L., Grman, M., Ondrias, K. & Ufnal, M. The impact of gut microbiota metabolites on cellular bioenergetics and cardiometabolic health. Nutr. Metab. 18, 72 (2021).
Maier, L. et al. Extensive impact of non-antibiotic drugs on human gut bacteria. Nature 555, 623–628 (2018).
Zimmermann, M., Zimmermann-Kogadeeva, M., Wegmann, R. & Goodman, A. L. Mapping human microbiome drug metabolism by gut bacteria and their genes. Nature 570, 462–467 (2019).
Forslund, K. et al. Disentangling type 2 diabetes and metformin treatment signatures in the human gut microbiota. Nature 528, 262–266 (2015).
Wu, H. et al. Metformin alters the gut microbiome of individuals with treatment-naive type 2 diabetes, contributing to the therapeutic effects of the drug. Nat. Med. 23, 850–858 (2017).
Sun, L. et al. Gut microbiota and intestinal FXR mediate the clinical benefits of metformin. Nat. Med. 24, 1919–1929 (2018).
Vieira-Silva, S. et al. Statin therapy is associated with lower prevalence of gut microbiota dysbiosis. Nature 581, 310–315 (2020).
Wilmanski, T. et al. Heterogeneity in statin responses explained by variation in the human gut microbiome. Med 3, 388–405.e6 (2022).
Klünemann, M. et al. Bioaccumulation of therapeutic drugs by human gut bacteria. Nature 597, 533–538 (2021).
Haiser, H. J. et al. Predicting and manipulating cardiac drug inactivation by the human gut bacterium Eggerthella lenta. Science 341, 295–298 (2013).
Maini Rekdal, V., Bess, E. N., Bisanz, J. E., Turnbaugh, P. J. & Balskus, E. P. Discovery and inhibition of an interspecies gut bacterial pathway for Levodopa metabolism. Science 364, eaau6323 (2019).
Zimmermann, M., Raosaheb Patil, K., Typas, A. & Maier, L. Towards a mechanistic understanding of reciprocal drug–microbiome interactions. Mol. Syst. Biol. 17, e10116 (2021).
Maier, L. & Typas, A. Systematically investigating the impact of medication on the gut microbiome. Curr. Opin. Microbiol. 39, 128–135 (2017).
Huang, S., Chaudhary, K. & Garmire, L. X. More is better: recent progress in multi-omics data integration methods. Front. Genet. 8, 84 (2017).
Bar, N. et al. A reference map of potential determinants for the human serum metabolome. Nature 588, 135–140 (2020).
Zeevi, D. et al. Personalized nutrition by prediction of glycemic responses. Cell 163, 1079–1094 (2015).
Berry, S. E. et al. Human postprandial responses to food and potential for precision nutrition. Nat. Med. 26, 964–973 (2020).
Doust, C. et al. Discovery of 42 genome-wide significant loci associated with dyslexia. Nat. Genet. 54, 1621–1629 (2022).
Gibson, G. R. et al. Dietary prebiotics: current status and new definition. Food Sci. Technol. Bull. 7, 1–19 (2010).
Hill, C. et al. Expert consensus document. The International Scientific Association for Probiotics and Prebiotics consensus statement on the scope and appropriate use of the term probiotic. Nat. Rev. Gastroenterol. Hepatol. 11, 506–514 (2014).
Swanson, K. S. et al. The International Scientific Association for Probiotics and Prebiotics (ISAPP) consensus statement on the definition and scope of synbiotics. Nat. Rev. Gastroenterol. Hepatol. 17, 687–701 (2020).
Salminen, S. et al. The International Scientific Association of Probiotics and Prebiotics (ISAPP) consensus statement on the definition and scope of postbiotics. Nat. Rev. Gastroenterol. Hepatol. 18, 649–667 (2021).
O’Toole, P. W., Marchesi, J. R. & Hill, C. Next-generation probiotics: the spectrum from probiotics to live biotherapeutics. Nat. Microbiol. 2, 17057 (2017).
Karcher, N. et al. Genomic diversity and ecology of human-associated Akkermansia species in the gut microbiome revealed by extensive metagenomic assembly. Genome Biol. 22, 209 (2021).
Depommier, C. et al. Supplementation with Akkermansia muciniphila in overweight and obese human volunteers: a proof-of-concept exploratory study. Nat. Med. 25, 1096–1103 (2019).
De Filippis, F., Esposito, A. & Ercolini, D. Outlook on next-generation probiotics from the human gut. Cell. Mol. Life Sci. 79, 76 (2022).
Baxter, M. & Colville, A. Adverse events in faecal microbiota transplant: a review of the literature. J. Hosp. Infect. 92, 117–127 (2016).
Maida, M., Mcilroy, J., Ianiro, G. & Cammarota, G. Faecal microbiota transplantation as emerging treatment in European countries. Adv. Exp. Med. Biol. 1050, 177–195 (2018).
Baunwall, S. M. D. et al. Danish national guideline for the treatment of infection and use of faecal microbiota transplantation (FMT). Scand. J. Gastroenterol. 56, 1056–1077 (2021).
Suskind, D. L. et al. Fecal microbial transplant effect on clinical outcomes and fecal microbiome in active Crohn’s disease. Inflamm. Bowel Dis. 21, 556–563 (2015).
Davar, D. et al. Fecal microbiota transplant overcomes resistance to anti–PD-1 therapy in melanoma patients. Science 371, 595–602 (2021).
Baruch, E. N. et al. Fecal microbiota transplant promotes response in immunotherapy-refractory melanoma patients. Science 371, 602–609 (2021).
Koopen, A. M. et al. Effect of fecal microbiota transplantation combined with mediterranean diet on insulin sensitivity in subjects with metabolic syndrome. Front. Microbiol. 12, 662159 (2021).
Ianiro, G. et al. Variability of strain engraftment and predictability of microbiome composition after fecal microbiota transplantation across different diseases. Nat. Med. 28, 1913–1923 (2022).
Valles-Colomer, M. et al. The person-to-person transmission landscape of the gut and oral microbiomes. Nature 614, 125–135 (2023).
Finlay, B. B., CIFAR Humans & The Microbiome. Are noncommunicable diseases communicable? Science 367, 250–251 (2020).
Aasmets, O., Krigul, K. L., Lüll, K., Metspalu, A. & Org, E. Gut metagenome associations with extensive digital health data in a volunteer-based Estonian microbiome cohort. Nat. Commun. 13, 869 (2022).
Gacesa, R. et al. Environmental factors shaping the gut microbiome in a Dutch population. Nature 604, 732–739 (2022).
Acknowledgements
This work was supported by the European Research Council (ERC-STG project MetaPG-716575 and ERC-CoG microTOUCH-101045015) to N.S. and by EMBO ALTF 593-2020 to M.V.-C. The work was also partially supported by the European Union’s Horizon 2020 program (ONCOBIOME-825410 project, MASTER-818368 project and IHMCSA-964590) to N.S., the European Union NextGenerationEU (Interconnected Nord-Est Innovation program, INEST) to N.S., the National Cancer Institute of the National Institutes of Health (1U01CA230551) to N.S. and the Premio Internazionale Lombardia e Ricerca 2019 to N.S. C.M. is funded by the Chronic Disease Research Foundation.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
S.E.B., A.M.V., T.D.S. and N.S. are consultants to Zoe Global. N.S. reports consultancy and/or Scientific Advisory Board contracts with Roche, YSOPIA Bioscience, Freya Biosciences and Alia Therapeutics and speaker fees from Illumina and is cofounder of PreBiomics. The other authors declare no competing interests.
Peer review
Peer review information
Nature Medicine thanks the anonymous reviewers for their contribution to the peer review of this work. Primary Handling Editor: Karen O’Leary, in collaboration with the Nature Medicine team.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary Table 1
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Valles-Colomer, M., Menni, C., Berry, S.E. et al. Cardiometabolic health, diet and the gut microbiome: a meta-omics perspective. Nat Med 29, 551–561 (2023). https://doi.org/10.1038/s41591-023-02260-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41591-023-02260-4