Abstract
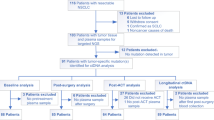
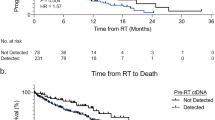
Circulating tumor DNA (ctDNA) sequencing guides therapy decisions but has been studied mostly in small cohorts without sufficient follow-up to determine its influence on overall survival. We prospectively followed an international cohort of 1,127 patients with non-small-cell lung cancer and ctDNA-guided therapy. ctDNA detection was associated with shorter survival (hazard ratio (HR), 2.05; 95% confidence interval (CI), 1.74–2.42; P < 0.001) independently of clinicopathologic features and metabolic tumor volume. Among the 722 (64%) patients with detectable ctDNA, 255 (23%) matched to targeted therapy by ctDNA sequencing had longer survival than those not treated with targeted therapy (HR, 0.63; 95% CI, 0.52–0.76; P < 0.001). Genomic alterations in ctDNA not detected by time-matched tissue sequencing were found in 25% of the patients. These ctDNA-only alterations disproportionately featured subclonal drivers of resistance, including RICTOR and PIK3CA alterations, and were associated with short survival. Minimally invasive ctDNA profiling can identify heterogeneous drivers not captured in tissue sequencing and expand community access to life-prolonging therapy.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Data availability
Genomic and clinical data are available on cBioPortal (https://www.cbioportal.org/study/summary?id=nsclc_ctdx_msk_2022). The raw sequencing data for MSK-IMPACT and MSK-ACCESS are protected and are not broadly available due to privacy laws. Researchers at MSK with appropriate IRB permission may request the data from the Center for Molecular Oncology (skicmopm@mskcc.org).
Code availability
No software was used for data collection.
References
Kris, M. G. et al. Using multiplexed assays of oncogenic drivers in lung cancers to select targeted drugs. JAMA 311, 1998–2006 (2014).
Howlader, N. et al. The effect of advances in lung-cancer treatment on population mortality. N. Engl. J. Med. 383, 640–649 (2020).
Pasche, B. & Grant, S. C. Non-small cell lung cancer and precision medicine a model for the incorporation of genomic features into clinical trial design. JAMA 311, 1975 (2014).
Bruno, D. S. et al. Racial disparities in biomarker testing and clinical trial enrollment in non-small cell lung cancer (NSCLC). J. Clin. Oncol. 39, 9005 (2021).
Robert, N. J. et al. Biomarker tissue journey among patients (pts) with untreated metastatic non-small cell lung cancer (mNSCLC) in the U.S. Oncology Network community practices. J. Clin. Oncol. 39, 9004 (2021).
Zugazagoitia, J. et al. Clinical utility of plasma-based digital next-generation sequencing in patients with advance-stage lung adenocarcinomas with insufficient tumor samples for tissue genotyping. Ann. Oncol. 30, 290–296 (2019).
Sheridan, C. Investors keep the faith in cancer liquid biopsies. Nat. Biotechnol. 37, 972–974 (2019).
Rolfo, C. et al. Liquid biopsy for advanced non-small cell lung cancer: a consensus statement from the International Association for the Study of Lung Cancer (IASLC). J. Thorac. Oncol. 16, 1647–1662 (2021).
Heitzer, E., Haque, I. S., Roberts, C. E. S. & Speicher, M. R. Current and future perspectives of liquid biopsies in genomics-driven oncology. Nat. Rev. Genet. 20, 71–88 (2019).
Sabari, J. K. et al. A prosspective study of circulating tumor DNA to guide matched targeted therapy in lung cancers. J. Natl Cancer Inst. 111, 575–583 (2019).
Oxnard, G. R. et al. Association between plasma genotyping and outcomes of treatment with osimertinib (AZD9291) in advanced non-small-cell lung cancer. J. Clin. Oncol. 34, 3375–3382 (2016).
Aggarwal, C. et al. Clinical implications of plasma-based genotyping with the delivery of personalized therapy in metastatic non-small cell lung cancer. JAMA Oncol. 5, 173 (2019).
Sacher, A. G. et al. Prospective validation of rapid plasma genotyping for the detection of EGFR and KRAS mutations in advanced lung cancer. JAMA Oncol. 2, 1014–1022 (2016).
Bettegowda, C. et al. Detection of circulating tumor DNA in early- and late-stage human malignancies. Sci. Transl. Med. 6, 224 (2014).
Leighl, N. B. et al. Clinical utility of comprehensive cell-free DNA analysis to identify genomic biomarkers in patients with newly diagnosed metastatic non-small cell lung cancer. Clin. Cancer Res. 25, 4691–4700 (2019).
Remon, J. et al. Osimertinib benefit in EGFR-mutant NSCLC patients with T790M-mutation detected by circulating tumour DNA. Ann. Oncol. 28, 784–790 (2017).
Tran, H. T. et al. Clinical outcomes in non-small cell lung cancer patients treated with EGFR-tyrosine kinase inhibitors and other targeted therapies based on tumor versus plasma genomic profiling. JCO Precis. Oncol. 5, 1241–1249 (2021).
Yang, M. et al. Incorporating blood-based liquid biopsy information into cancer staging: time for a TNMB system? Ann. Oncol. 29, 311–323 (2018).
Chaudhuri, A. A. et al. Early detection of molecular residual disease in localized lung cancer by circulating tumor DNA profiling. Cancer Discov. 7, 1394–1403 (2017).
Nabet, B. Y. et al. Noninvasive early identification of therapeutic benefit from immune checkpoint inhibition. Cell 183, 363–376 (2020).
Goldberg, S. B. et al. Early assessment of lung cancer immunotherapy response via circulating tumor DNA. Clin. Cancer Res. 24, 1872–1880 (2018).
Gandara, D. R. et al. Blood-based tumor mutational burden as a predictor of clinical benefit in non-small cell lung cancer patients treated with atezolizumab. Nat. Med. 24, 1441–1448 (2018).
Karachaliou, N. et al. Association of EGFR L858R mutation in circulating free DNA with survival in the EURTAC trial. JAMA Oncol. 1, 149–157 (2015).
Mack, P. C. et al. Spectrum of driver mutations and clinical impact of circulating tumor DNA analysis in non-small cell lung cancer: analysis of over 8000 cases. Cancer 126, 3219–3228 (2020).
Newman, A. M. et al. Integrated digital error suppression for improved detection of circulating tumor DNA. Nat. Biotechnol. 34, 547–555 (2016).
Razavi, P. et al. High-intensity sequencing reveals the sources of plasma circulating cell-free DNA variants. Nat. Med. 25, 1928–1937 (2019).
Abbosh, C. et al. Phylogenetic ctDNA analysis depicts early-stage lung cancer evolution. Nature 545, 446–451 (2017).
Merker, J. D. et al. Circulating tumor DNA analysis in patients with cancer: American Society of Clinical Oncology and College of American Pathologists joint review. J. Clin. Oncol. 36, 1631–1641 (2018).
Alix-Panabières, C. & Pantel, K. Liquid biopsy: from discovery to clinical application. Cancer Discov. 11, 858–873 (2021).
Paweletz, C. P. et al. Bias-corrected targeted next-generation sequencing for rapid, multiplexed detection of actionable alterations in cell-free DNA from advanced lung cancer patients. Clin. Cancer Res. 22, 915–922 (2016).
Supplee, J. G. et al. Sensitivity of next-generation sequencing assays detecting oncogenic fusions in plasma cell-free DNA. Lung Cancer 134, 96–99 (2019).
Chakravarty, D. et al. OncoKB: a precision oncology knowledge base. JCO Precis. Oncol. 2017, 1–16 (2017).
Mandelker, D. et al. Mutation detection in patients with advanced cancer by universal sequencing of cancer-related genes in tumor and normal DNA vs guideline-based germline testing. JAMA 318, 825–835 (2017).
Zehir, A. et al. Mutational landscape of metastatic cancer revealed from prospective clinical sequencing of 10,000 patients. Nat. Med. 23, 703–713 (2017).
Brannon, A. R. et al. Enhanced specificity of high sensitivity somatic variant profiling in cell-free DNA via paired normal sequencing: design, validation, and clinical experience of the MSK-ACCESS liquid biopsy assay. Nat. Comm. 12, 3770 (2021).
Li, B. T. et al. Ultra-deep next-generation sequencing of plasma cell-free DNA in patients with advanced lung cancers: results from the Actionable Genome Consortium. Ann. Oncol. J. Eur. Soc. Med. Oncol. 30, 597–603 (2019).
Nakamura, Y. et al. Clinical utility of circulating tumor DNA sequencing in advanced gastrointestinal cancer: SCRUM-Japan GI-SCREEN and GOZILA studies. Nat. Med. 26, 1859–1864 (2020).
Pairawan, S. et al. Cell-free circulating tumor DNA variant allele frequency associates with survival in metastatic cancer. Clin. Cancer Res. 26, 1924–1931 (2020).
Ramalingam, S. S. et al. Abstract CT078: tumor mutational burden (TMB) as a biomarker for clinical benefit from dual immune checkpoint blockade with nivolumab (nivo) + ipilimumab (ipi) in first-line (1L) non-small cell lung cancer (NSCLC): identification of TMB cutoff from CheckMate 568. Cancer Research 78, CT078 (2018).
Chabon, J. J. et al. Integrating genomic features for non-invasive early lung cancer detection. Nature 580, 245–251 (2020).
Morbelli, S. et al. Circulating tumor DNA reflects tumor metabolism rather than tumor burden in chemotherapy-naive patients with advanced non-small cell lung cancer (NSCLC): 18F-FDG PET/CT study. J. Nucl. Med. 58, 1764–1769 (2017).
Piotrowska, Z. et al. Landscape of acquired resistance to osimertinib in EGFR-mutant NSCLC and clinical validation of combined EGFR and RET inhibition with osimertinib and BLU-667 for acquired RET fusion. Cancer Discov. 8, 1529–1539 (2018).
Offin, M. et al. Acquired ALK and RET gene fusions as mechanisms of resistance to osimertinib in EGFR-mutant lung cancers. JCO Precis. Oncol. 2, PO.18.00126.
Piper-Vallillo, A. J., Sequist, L. V. & Piotrowska, Z. Emerging treatment paradigms for EGFR-mutant lung cancers progressing on osimertinib: a review. J. Clin. Oncol. 38, 2926–2936 (2020).
Tan, A. C. Targeting the PI3K/Akt/mTOR pathway in non-small cell lung cancer (NSCLC). Thorac. Cancer 11, 511–518 (2020).
Nicholas, M. et al. Clonal status of actionable driver events and the timing of mutational processes in cancer evolution. Sci. Transl. Med. 7, 283ra54 (2015).
Shen, R. et al. Harnessing clinical sequencing data for survival stratification of patients with metastatic lung adenocarcinomas. JCO Precis. Oncol. https://doi.org/10.1200/po.18.00307 (2019).
Bolton, K. L. et al. Cancer therapy shapes the fitness landscape of clonal hematopoiesis. Nat. Genet. 52, 1219–1226 (2020).
Coombs, C. C. et al. Therapy-related clonal hematopoiesis in patients with non-hematologic cancers is common and associated with adverse clinical outcomes. Cell Stem Cell 21, 374–382 (2017).
Bolton, K. L. et al. Clonal hematopoiesis is associated with risk of severe Covid-19. Nat. Commun. 12, 5975 (2021).
Ignatiadis, M., Sledge, G. W. & Jeffrey, S. S. Liquid biopsy enters the clinic – implementation issues and future challenges. Nat. Rev. Clin. Oncol. 18, 297–312 (2021).
Lisberg, A. et al. A phase II study of pembrolizumab in EGFR-mutant, PD-L1+, tyrosine kinase inhibitor (TKI) naive patients with advanced NSCLC. J. Thorac. Oncol. 13, 1138–1145 (2018).
Cristiano, S. et al. Genome-wide cell-free DNA fragmentation in patients with cancer. Nature 570, 385–389 (2019).
Parikh, A. R. et al. Liquid versus tissue biopsy for detecting acquired resistance and tumor heterogeneity in gastrointestinal cancers. Nat. Med. 25, 1415–1421 (2019).
Jamal-Hanjani, M. et al. Tracking the evolution of non-small cell lung cancer. N. Engl. J. Med. 376, 2109–2121 (2017).
Mateo, J. et al. A framework to rank genomic alterations as targets for cancer precision medicine: the ESMO Scale for Clinical Actionability of Molecular Targets (ESCAT). Ann. Oncol. J. Eur. Soc. Med. Oncol. 29, 1895–1902 (2018).
Ramalingam, S. S. et al. Overall survival with osimertinib in untreated, EGFR-mutated advanced NSCLC. N. Engl. J. Med. 382, 41–50 (2020).
Newman, A. M. et al. An ultrasensitive method for quantitating circulating tumor DNA with broad patient coverage. Nat. Med. 20, 548–554 (2014).
Presley, C. J. et al. Association of broad-based genomic sequencing with survival among patients with advanced non-small cell lung cancer in the community oncology setting. JAMA 320, 469–477 (2018).
Rothwell, D. G. et al. Utility of ctDNA to support patient selection for early phase clinical trials: the TARGET study. Nat. Med. 25, 738–743 (2019).
Murthy, V. H., Krumholz, H. M. & Gross, C. P. Participation in cancer clinical trials: race-, sex-, and age-based disparities. JAMA 291, 2720–2726 (2004).
Scher, K. S. & Hurria, A. Under-representation of older adults in cancer registration trials: known problem, little progress. J. Clin. Oncol. 30, 2036–2038 (2012).
Sedrak, M. S. et al. Older adult participation in cancer clinical trials: a systematic review of barriers and interventions. CA Cancer J. Clin. 71, 78–92 (2021).
Rolfo, C., Russo, A. & de Miguel-Pérez, D. Speeding tumor genotyping during the SARS-CoV-2 outbreak through liquid biopsy. Cancer 126, 4089–4091 (2020).
Li, B. T. et al. Reimagining patient-centric cancer clinical trials: a multi-stakeholder international coalition. Nat. Med. 28, 620–626 (2022).
Syeda, M. M. et al. Circulating tumour DNA in patients with advanced melanoma treated with dabrafenib or dabrafenib plus trametinib: a clinical validation study. Lancet Oncol. 22, 370–380 (2021).
Turner, N. C. et al. Circulating tumour DNA analysis to direct therapy in advanced breast cancer (plasmaMATCH): a multicentre, multicohort, phase 2a, platform trial. Lancet Oncol. 21, 1296–1308 (2020).
Chabon, J. J. et al. Circulating tumour DNA profiling reveals heterogeneity of EGFR inhibitor resistance mechanisms in lung cancer patients. Nat. Commun. 7, 11815 (2016).
Bardelli, A. & Pantel, K. Liquid biopsies, what we do not know (yet). Cancer Cell. 31, 172–179 (2017).
Siravegna, G. et al. Clonal evolution and resistance to EGFR blockade in the blood of colorectal cancer patients. Nat. Med. 21, 795–801 (2015).
Lennon, A. M. et al. Feasibility of blood testing combined with PET-CT to screen for cancer and guide intervention. Science 369, 6499 (2020).
Karczewski, K. J. et al. The mutational constraint spectrum quantified from variation in 141,456 humans. Nature 581, 434–443 (2020).
McNulty, S. N., Parikh, B. A., Duncavage, E. J., Heusel, J. W. & Pfeifer, J. D. Optimization of population frequency cutoffs for filtering common germline polymorphisms from tumor-only next-generation sequencing data. J. Mol. Diagn. 21, 903–912 (2019).
Shen, R. & Seshan, V. E. FACETS: allele-specific copy number and clonal heterogeneity analysis tool for high-throughput DNA sequencing. Nucleic Acids Res. 44, 16 (2016).
Kanoun, S. et al. Influence of software tool and methodological aspects of total metabolic tumor volume calculation on baseline [18F]FDG PET to predict survival in Hodgkin lymphoma. PLoS ONE 10, e0140830 (2015).
Boellaard, R. et al. FDG PET and PET/CT: EANM procedure guidelines for tumour PET imaging: version 1.0. Eur. J. Nucl. Med. Mol. Imaging 37, 181 (2009).
Acknowledgements
This work was supported by a grant from the Antidote Health Foundation for Cure of Cancer (BTL), the National Institutes of Health (T32-CA009207 (J.J.), CTSA UL1TR00457 (M.R.M.-G.), P50 CA247749 01 and P30 CA008748), the Molecular Diagnostics Service in the Department of Pathology, and the Marie-Josee and Henry R. Kravis Center for Molecular Oncology.
Author information
Authors and Affiliations
Contributions
Conception: J.J. and B.T.L. Radiomic analysis: J.J., E.S.L., R.Y. and J.P.D. Patient accrual: J.J., A.N., P.K.P., J.E.C., Y.R.M.-G., B.D., H.A.Y., M.O., M.D.H., K.C.A., M.G.Z., M.G.K., K.K.N., J.E., I.P., W.V.L., J.J.F., A.I., D.M., G.R., B.J.P., D.L.C., C.I.D., M.I., S.C., N.P., A.L., N.R., J.C., W.D.T., G.J.R., V.W.R., A.R., D.G., A.D., D.R.J., C.M.R., J.M.I. and B.T.L. Genomic data collection and analysis: J.J., G.J., A.R.B., R.B., A.Z., M.D., N.S., D.C., R.K., R.M., S.P.S., M.F.B., M.E.A., M.L., R.L., L.P.L. and M.L. Clinical data collection and analysis: J.J., E.S.L., N.S., Y.R.M.-G., H.-Y.T., C.-R.X., C.T.-L. and M.D.S. Administration: A.M., J.G., D.B.S., A.D., H.I.S., P.L., L.P.L., M.F.B., M.E.A., M.L., P.R., J.S.R.-F., D.R.J., C.M.R., J.M.I. and B.T.L. Statistical plan: J.J., M.G., R.S. and S.P.S. Writing: J.J., C.W., P.R., J.S.R.-F. and B.T.L. All co-authors reviewed and approved the final draft of the manuscript.
Corresponding author
Ethics declarations
Competing interests
J.J. has a patent licensed by MDSeq. A.N. reports serving as a one-time paid consultant for Bayer. P.K.P. receives compensation for consulting or advisory board participation from Bicara Therapeutics, Boehringer Ingelheim, EMD Serono, GlaxoSmithKline, Takeda Pharmaceuticals, WC Communications and Xencor and receives honoraria for participation in CME educational programs from PeerVoice, ACE Oncology Research to Practice, Clinical Care Options, Spring to Life and Touch Independent Medical Education. J.E.C. has served as a consultant for Astra Zeneca, Bristol-Myers Squib, Genentech, Merck, Flame Biosciences, Novartis, Regeneron-Sanofi, Guardant Health and Janssen and has received research funding to her institution from Astra Zeneca, Bristol-Myers Squib, Genentech and Merck. A.R.B. has stock ownership in Johnson & Johnson. R.B. reports a grant from Archer, honoraria for advisory board participation from Loxo oncology and speaking fees from Illumina. A.Z. has received speaking fees from Illumina. D.C. has consulted with/received honoraria from Pfizer, Loxo/Lilly Oncology, BridgeBio, FORE Therapeutics, Scorpion Therapeutics and Vividion Therapeutics. Y.R.M.-G. acknowledges receipt of training through an institutional K30 grant from the NIH (CTSA UL1TR00457). She has received funding from a Kristina M. Day Young Investigator Award from Conquer Cancer, the ASCO Foundation, funded by Charles M. Baum and Carol A. Baum. She reports travel, accommodation and expenses from AstraZeneca and honoraria from Virology Education. She acknowledges research funding to the institution from Loxo Oncology at Eli Lilly, Elucida Oncology, Taiho Oncology, Hengrui USA/Jiangsu Hengrui Pharmaceuticals and Endeavor Biomedicines. She acknowledges royalties from Rutgers University Press and Wolters Kluwer. H.-Y.T. has received academic travel support from Resolution Bioscience. C.X. has received honoraria from AstraZeneca, BeiGene, Boehringer Ingelheim, Bristol Myers Squibb, Lilly, Merck & Co., Novartis, Pfizer and Roche; has received research support from BeiGene and Hengrui Pharmaceutical; and has received reimbursement for travel and accommodation expenses from AstraZeneca, Boehringer Ingelheim, Bristol Myers Squibb, Lilly, Merck & Co., Novartis, Pfizer and Roche. B.D. reports equity interest in Eli Lilly and Company, Roche and CVS Health and family member involvement with Eli Lilly and Company and CVS Health. H.A.Y. has consulted for AstraZeneca, Blueprint Medicine, Janssen Oncology, Cullinan and Daiichi. Her institution has received research funding for clinical trials from AstraZeneca, Daiichi, Pfizer, Novartis, Cullinan Oncology and Lilly. M.O. reports personal fees from PharmaMar, personal fees from Novartis, personal fees from Targeted Oncology, personal fees from Bristol-Myers Squibb, personal fees from Merck Sharp & Dohme, personal fees from Jazz Pharmaceuticals and personal fees from Astro Pharmaceuticals, outside the submitted work. M.D.H. reports grants from BMS and personal fees from Achilles, Adagene, Adicet, Arcus, AstraZeneca, Blueprint, BMS, DaVolterra, Eli Lilly, Genentech/Roche, Genzyme/Sanofi, Janssen, Immunai, Instil Bio, Mana Therapeutics, Merck, Mirati, Natera, Pact Pharma, Shattuck Labs and Regeneron, as well as equity options from Factorial, Immunai, Shattuck Labs and Arcus. A patent filed by Memorial Sloan Kettering related to the use of TMB to predict response to immunotherapy (PCT/US2015/062208) is pending and licensed by PGDx. P.L. is listed as an inventor on patent applications filed by MSKCC that describe approaches to treat KRAS or BRAF-mutant tumors. K.C.A. reports personal fees from AstraZeneca and nonfinancial support from Takeda and Novartis outside the submitted work. In the last 3 years, M.G.Z. has received consulting fees from Takeda, GlaxoSmithKline, Expert Connect, Aldeyra Therapeutics, Novocure and Atara and honoraria from Research to Practice, Medical Learning Institute and OncLive. Memorial Sloan Kettering receives research funding from the Department of Defense, the National Institutes of Health, Precog, GlaxoSmithKline, Epizyme, Polaris, Sellas Life Sciences, Bristol Myers Squibb, Millenium/Takeda, Curis and Atara for research conducted by M.G.Z. M.G.Z. serves as chair of the board of directors of the Mesothelioma Applied Research Foundation, uncompensated. M.G.K. receives personal fees from Novartis, Sanofi-Genzyme, AstraZeneca, Pfizer, Janssen and Daiichi-Sankyo; received honoraria for participation in educational programs from WebMD, OncLive, Physicians Education Resources, Prime Oncology, Intellisphere, Creative Educational Concepts, Peerview, i3 Health, Paradigm Medical Communications, AXIS, Carvive Systems and AstraZeneca; received travel support from AstraZeneca, Pfizer and Genentech; and received editorial support from Hoffman La-Roche. Memorial Sloan Kettering has received research funding from the National Cancer Institute (USA), the Lung Cancer Research Foundation and Genentech Roche for research conducted by M.G.K. I.P. has consulted or served on the advisory boards of Pfizer, AstraZeneca, Blueprint Medicine, DavaOncology, Eli Lilly and Curio Science. W.V.L. receives institutional research funding from Daiichi Sankyo, Amgen and Abbvie and has been a compensated consultant for PharmaMar, G1 Therapeutics, AstraZeneca and Jazz Pharmaceuticals. D.M. is a consultant for AstraZeneca, Johnson & Johnson, Boston Scientific, Bristol Myers Squibb and Merck. G.R. receives royalties from Scanlan International. B.J.P. is a consultant for AstraZeneca. L.P.L. is an employee and shareholder of Agilent Technologies. M.L. is an employee and shareholder of Agilent Technologies. C.I.D. reports serving on the advisory board for Amgen and Ipsen, honoraria for Merck and academic travel support from Roche. M.I. reports serving on the advisory boards of Pfizer and Takeda; receiving honoraria from Roche, AstraZeneca, MSD, Bristol Myers Squibb, Pfizer, Takeda and Novartis; and travel support from Roche, AstraZeneca and MSD. S.C. reports advisory board fees from Roche and Astra Zeneca and travel support from Bristol Myers Squibb, outside the submitted work. N.P. has served on advisory boards or received personal honoraria from Boehringer Ingelheim, MSD, Merck, Bristol-Myers Squib, Astra Zeneca, Takeda, Pfizer, Roche, Novartis, Ipsen and Bayer and has received research funding to his institution from Bayer, Pfizer and Roche. A.L. reports personal fees and travel funding from Mundipharma/Helsinn, personal fees from Bayer and personal fees from Eisai. G.J.R. reports grants from National Institutes of Health/National Cancer Institute, has been an uncompensated consultant to Daiichi, Pfizer and Mirati and has institutional research support from Mirati, Takeda, Merck, Roche, Pfizer and Novartis. In addition, G.J.R. has pending patents US20170273982A1 and WO2017164887A8. D.B.S. has served as a consultant for/received honoraria from Loxo Oncology, Lilly Oncology, Pfizer, QED Therapeutics, Vivideon Therapeutics and Illumina. M.L. has received honoraria for advisory board participation from Merck, Astra-Zeneca, Bristol Myers Squibb, Blueprint Medicines, Janssen Pharmaceuticals, Takeda Pharmaceuticals, Lilly Oncology, LOXO Oncology, Bayer, ADC Therapeutics, Riken Genesis and Paige AI and research support from LOXO Oncology, Merus, and Helsinn Therapeutics. Marc Ladanyi reports honoraria for ad-hoc advisory board participation from Merck, AstraZeneca, Bristol Myers Squibb, Takeda, Bayer, and Lilly Oncology; and research support from LOXO Oncology, Merus and Helsinn Therapeutics. V.W.R. reports grants from Genelux, grants from Genentech, other from DaVinci Surgery, nonfinancial support from Bristol Myers Squibb and personal fees from NIH/Coordinating Center for Clinical Trials, outside the submitted work. A.R. reports grants from Varian Medical Systems, grants and personal fees from AstraZeneca, grants and personal fees from Merck, grants and personal fees from Boehringer Ingelheim, grants from Pfizer, personal fees from Research to Practice, personal fees from Cybrexa, personal fees from More Health and nonfinancial support from Philips/Elekta, outside the submitted work. D.G. reports grants from Varian, AstraZeneca, Merck and Bristol Myers Squibb and personal fees from Varia, AstraZeneca, Merck, US Oncology, Bristol Myers Squibb, Relfexion, WebMD, Vindico and Medscape and has served on the advisory board for AstraZeneca. A.D. has received honoraria or worked on the advisory boards of Ignyta/Genentech/Roche, Loxo/Bayer/Lilly, Takeda/Ariad/Millenium, TP Therapeutics, AstraZeneca, Pfizer, Blueprint Medicines, Helsinn, Beigene, BergenBio, Hengrui Therapeutics, Exelixis, Tyra Biosciences, Verastem, MORE Health, Abbvie, 14ner/Elevation Oncology, Remedica, ArcherDX, Monopteros, Novartis, EMD Serono, Melendi, Liberum, Repare RX, Nuvalent, Merus, AXIS, Chugai Pharm and EPG Health; has received funding through his institution from Pfizer, Exelixis, GlaxoSmithKline, Teva, Taiho and PharmaMar; has received research support from Foundation Medicine; receives royalties from Wolters Kluwer; has received other support from Boehringer Ingelheim; has received food/beverage from Merck, Puma and Merus; and has received CME honoraria from Medscape, OncLive, PeerVoice, Physicians Education Resources, Targeted Oncology, Research to Practice, Axis, Peerview Institute, Paradigm Medical Communications, WebMD, MJH Life Sciences, Med Learning, Imedex, Answers in CME and Clinical Care Options. H.I.S. reports the following support: compensated consultant/advisor to Ambry Genetics, Konica Minolta, Bayer, Pfizer, Sun Pharmaceuticals and WCG Oncology; uncompensated consultant/advisory to Amgen, ESSA Pharma, Janssen Research & Development, Janssen Biotech and Sanofi Aventis; he has received research funding (to his institution) from Epic Sciences, Illumina, Janssen, Menarini Silicon Biosystems, Prostate Cancer Foundation and ThermoFisher; intellectual property rights from BioNTech, Elucida Oncology, MaBVAX and Y-mAbs Therapeutics; and nonfinancial support from Amgen, Asterias Biotherapeutics, Bayer, ESSA Pharma, Menarini Silicon Biosystems, Phosplatin, Pfizer, Prostate Cancer Foundation and WCG Oncology. S.P.S. is a shareholder and consultant of Canesia Health. M.F.B. reports a consulting/advisory role with PetDx and Eli Lilly; research support from Grail; and a patent pending related to cfDNA profiling. R.L. is on the supervisory board of Qiagen and is a scientific advisor to Imago, Mission Bio, Syndax. Zentalis, Ajax, Bakx, Auron, Prelude, C4 Therapeutics and Isoplexis for which he receives equity support. He receives research support from Ajax and Abbvie and has consulted for Incyte, Janssen, Morphosys and Novartis. He has received honoraria from Astra Zeneca and Kura for invited lectures and from Gilead for grant reviews. P.R. received institutional grant/funding from Grail, Illumina, Novartis, Epic Sciences and ArcherDx and consultation/ad board/honoraria from Novartis, Foundation Medicine, AstraZeneca, Epic Sciences, Inivata, Natera and Tempus. J.R.-F. is a paid consultant of Goldman Sachs, Paige.AI and REPARE Therapeutics, a member of the scientific advisory board of Goldman Sachs, Paige.AI and Volition RX, and an ad hoc member of the scientific advisory board of Roche, Genentech, Roche Tissue Diagnostics, Ventana, Novartis, InVicro and GRAIL. D.R.J. serves as a consultant for AstraZeneca and Merck. C.M.R. reports personal fees from AbbVie, Amgen, Ascentage, AstraZeneca, Bicycle, Celgene, Daiichi Sankyo, Genentech/Roche, Ipsen, Jansen, Jazz, Lilly/Loxo, Pfizer, PharmaMar, Syros, Vavotek, Bridge Medicines and Harpoon Therapeutics, outside the submitted work. J M.I. reports equity in LumaCyte and has served as an uncompensated member of a steering committee for Genentech. B.T.L. has served as an uncompensated advisor and consultant to Amgen, Genentech, Boehringer Ingelheim, Lilly, AstraZeneca and Daiichi Sankyo. He has received research grants to his institution from Amgen, Genentech, AstraZeneca, Daiichi Sankyo, Lilly, Illumina, GRAIL, Guardant Health, Hengrui Therapeutics, MORE Health and Bolt Biotherapeutics. He has received academic travel support from MORE Health and Jiangsu Hengrui Medicine. He is an inventor on two institutional patents at Memorial Sloan Kettering (US62/685,057, US62/514,661) and has intellectual property rights as a book author at Karger Publishers and Shanghai Jiao Tong University Press. The remaining authors declare no competing interests.
Peer review
Peer review information
Nature Medicine thanks Ana Vivancos, Benjamin Besse and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Primary Handling Editor: Joao Monteiro, in collaboration with the Nature Medicine team.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Turnaround time of plasma and tissue sequencing.
Boxplots showing median (center) +/−25%ile (boxes) and 95%ile (whiskers) of turnaround time for liquid biopsy (MSK-ACCESS and ctDx Lung, N independent samples = 2,162) and tissue (MSK-IMPACT, N independent samples = 612) sequencing from date of blood collection. Tissue start time is the date of white blood cell control collection. The turnaround time for plasma ctDNA sequencing was significantly faster than for tissue sequencing (*two-sided Mann-Whitney U, p < 0.001).
Extended Data Fig. 2 Correlates of ctDNA alteration levels.
Histograms showing the proportion of patients with either number of ctDNA alterations or maximum mutation variant allele frequency (VAF) per sample. P-values are from Mann-Whitney U tests or Kruskal-Wallis tests for histologic subgroups. Raw VAFs of zero are set to the minimum value of the log axis.
Extended Data Fig. 3 Survival of patients without tissue sequencing matched to targeted therapy by ctDNA.
Kaplan-Meier survival curves for patients without tissue sequencing matched or not matched to targeted therapy. Number at risk in each category is adjusted for left truncation and time-dependent nature of targeted therapy variables.
Supplementary information
Supplementary Information
Supplemental text and Supplementary Figs. S1–14.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Jee, J., Lebow, E.S., Yeh, R. et al. Overall survival with circulating tumor DNA-guided therapy in advanced non-small-cell lung cancer. Nat Med 28, 2353–2363 (2022). https://doi.org/10.1038/s41591-022-02047-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41591-022-02047-z
This article is cited by
-
HER2-targeted therapies in cancer: a systematic review
Biomarker Research (2024)
-
Oncogenic alterations in advanced NSCLC: a molecular super-highway
Biomarker Research (2024)
-
Enhancing NSCLC recurrence prediction with PET/CT habitat imaging, ctDNA, and integrative radiogenomics-blood insights
Nature Communications (2024)
-
Associations between detectable circulating tumor DNA and tumor glucose uptake measured by 18F-FDG PET/CT in early-stage non-small cell lung cancer
BMC Cancer (2023)
-
Prognostic value of circulating tumor DNA in operable non-small cell lung cancer: a systematic review and reconstructed individual patient-data based meta-analysis
BMC Medicine (2023)